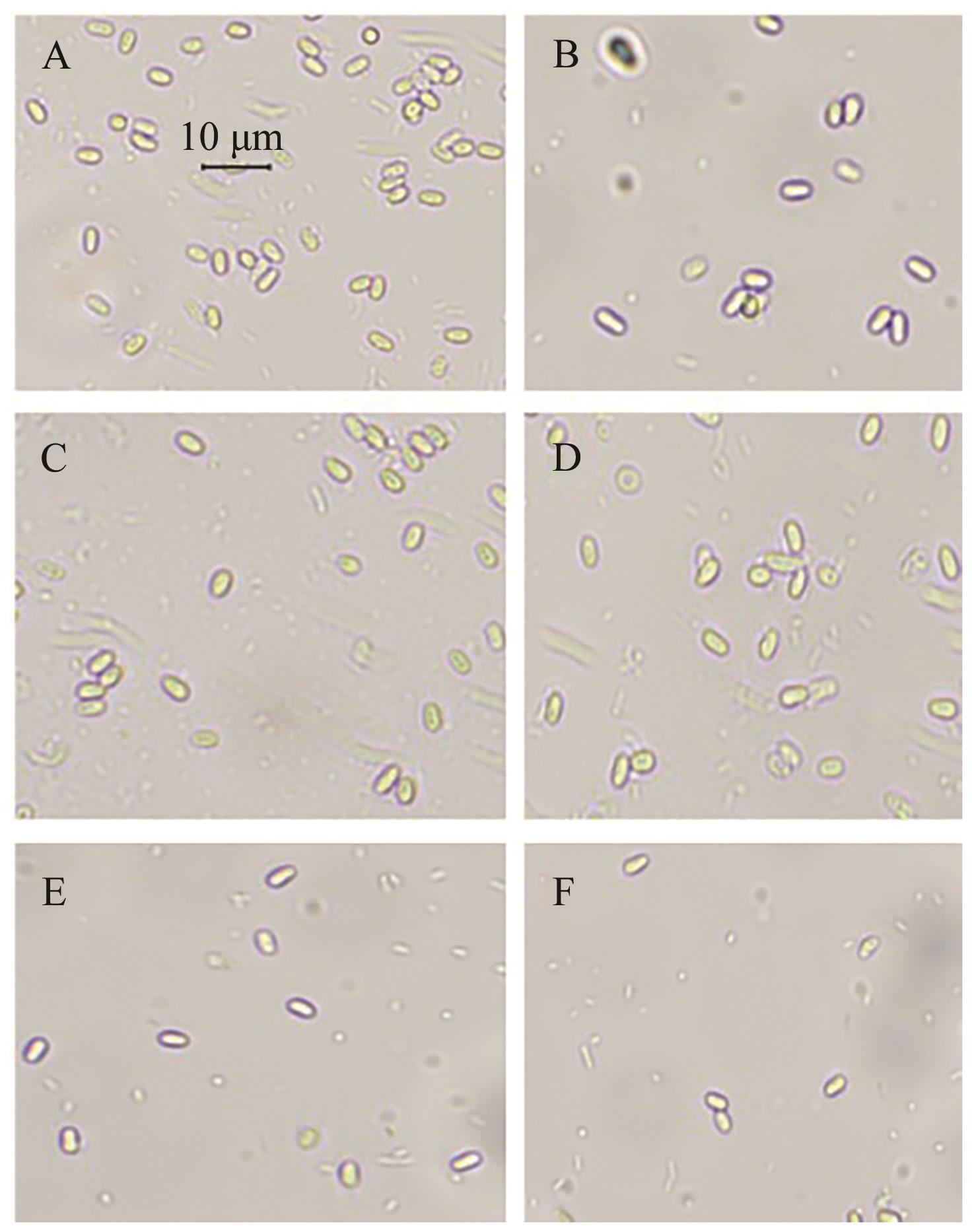
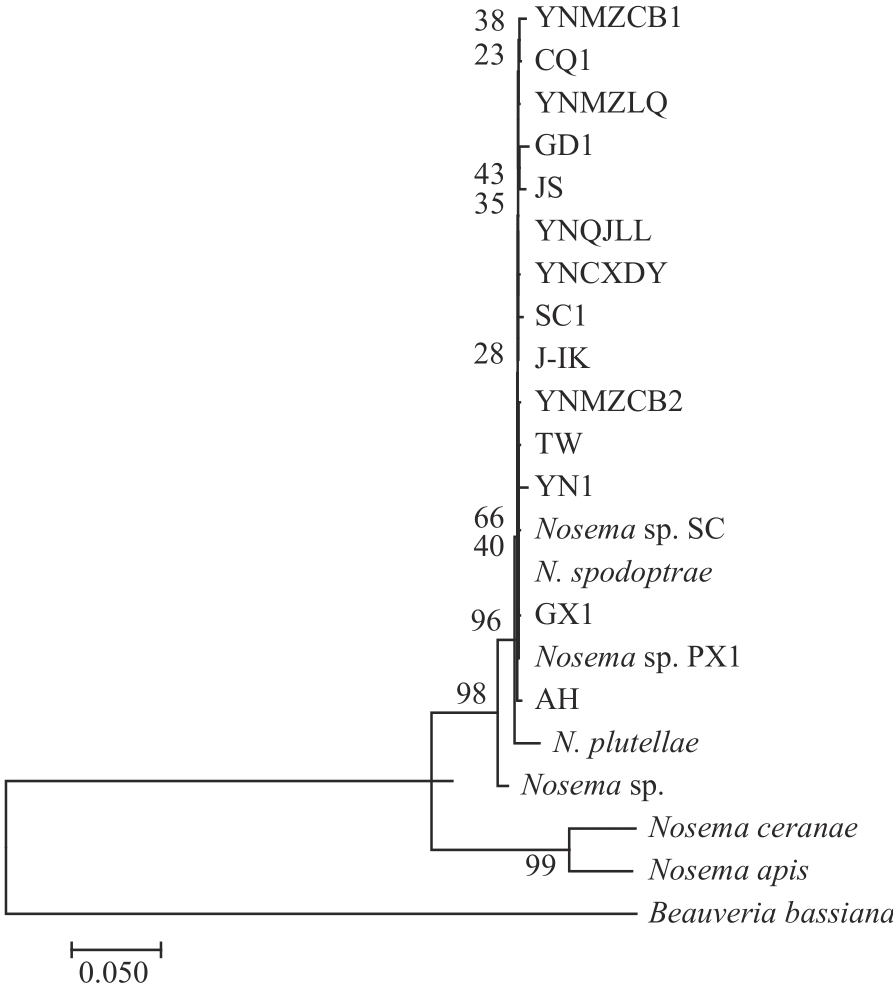
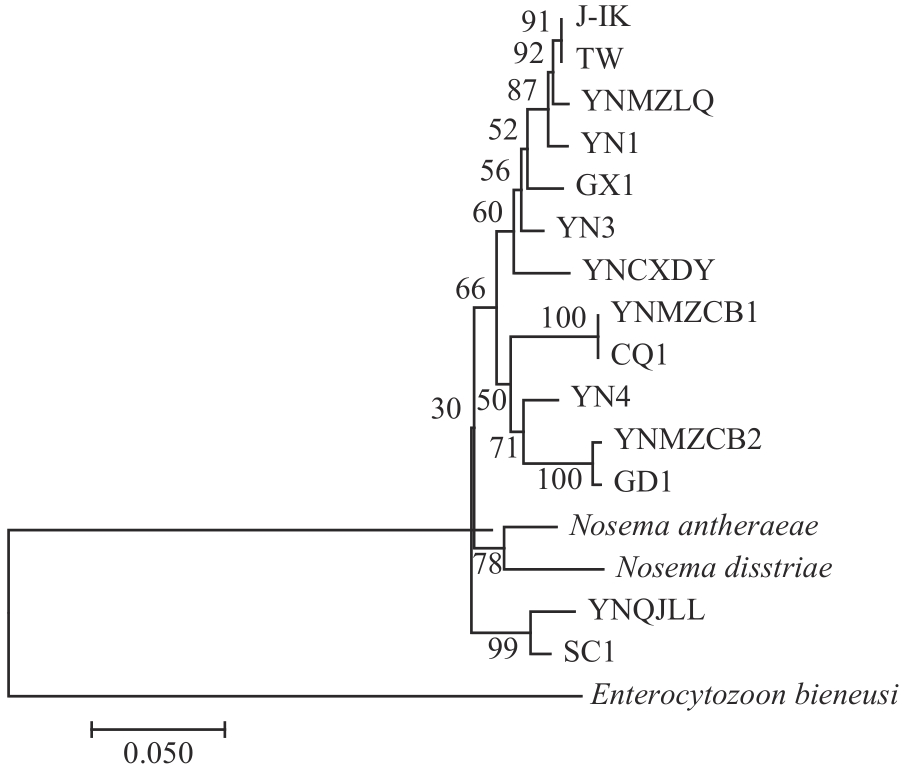
Current Biotechnology ›› 2024, Vol. 14 ›› Issue (4): 631-639.DOI: 10.19586/j.2095-2341.2024.0019
• Articles • Previous Articles Next Articles
Analysis of Genetic Diversity of Nosema bombycis in Yunnan Sericulture Region
Yonghong ZHANG1( ), Zhenguo SU1, Jiafu LUO1, Chunfeng LI2(
), Zhenguo SU1, Jiafu LUO1, Chunfeng LI2( ), Baolin AO3
), Baolin AO3
- 1.Institute of Sericulture and Apiculture,Yunnan Academy of Agricultural Science,Yunnan Mengzi 661101,China
2.Chongqing Key Laboratory of Microsporidia Infection and Control,Southwest University,Chongqing 400715,China
3.Mulberry-silkworm Service Center of Luliang County,Yunnan Luliang 655600,China
-
Received:2024-02-04Accepted:2024-04-24Online:2024-07-25Published:2024-08-07 -
Contact:Chunfeng LI
云南蚕区家蚕微孢子虫遗传多样性分析
张永红1( ), 苏振国1, 罗家福1, 李春峰2(
), 苏振国1, 罗家福1, 李春峰2( ), 敖宝林3
), 敖宝林3
- 1.云南省农业科学院蚕桑蜜蜂研究所,云南 蒙自 661101
2.西南大学,微孢子虫感染与防控重庆市重点实验室,重庆 400715
3.陆良县蚕桑服务中心,云南 陆良 655600
-
通讯作者:李春峰 -
作者简介:张永红 E-mail: zhang200503@126.com; -
基金资助:云南省农业科学院预研项目(2024KYZX-01);重庆市重点实验室开放课题项目(CKMIC202102)
CLC Number:
Cite this article
Yonghong ZHANG, Zhenguo SU, Jiafu LUO, Chunfeng LI, Baolin AO. Analysis of Genetic Diversity of Nosema bombycis in Yunnan Sericulture Region[J]. Current Biotechnology, 2024, 14(4): 631-639.
张永红, 苏振国, 罗家福, 李春峰, 敖宝林. 云南蚕区家蚕微孢子虫遗传多样性分析[J]. 生物技术进展, 2024, 14(4): 631-639.
share this article
| 名称 | 宿主 | SSU rDNA GenBank登录号 | ITS GenBank登录号 | 地理来源 |
|---|---|---|---|---|
| YNMZCB1 | Bombyx mori | OQ970549 | OQ974967 | 云南省蒙自市 |
| YNMZCB2 | OQ970550 | OQ974968 | 云南省蒙自市 | |
| YNMZLQ | OQ970551 | OQ974969 | 云南省蒙自市 | |
| YNCXDY | OR597059 | OR597057 | 云南省大姚县 | |
| YNQJLL | OR597056 | OR597058 | 云南省陆良县 | |
| YN1 | JF334596 | JF443647 | 云南省 | |
| YN2 | JF334597 | JF443648 | 云南省 | |
| YN3 | JF334598 | JF443649 | 云南省 | |
| YN4 | JF334599 | JF443650 | 云南省 | |
| SC1 | JF334592 | JF443631 | 四川省 | |
| CQ1 | EU350391 | EU350394 | 重庆市 | |
| GX1 | JF443547 | JF443600 | 广西壮族自治区 | |
| GD1 | JF334582 | JF443619 | 广东省 | |
| AH | GQ334399 | — | 安徽省 | |
| JS | AY616662 | — | 江苏省 | |
| TW | AY209011 | AY209011 | 台湾省 | |
| J-IK | AY259631 | AY259631 | 日本茨城县 | |
| Nosema ceranae | Apis mellifera | DQ486027 | — | 台湾省 |
| Nosema apis | Apis cerana | U97150 | — | 美国加利福尼亚州 |
| Nosema sp.SC | Samia cynthia ricini | FJ767862 | — | 江苏省 |
| Nosema sp.PX1 | Plutella xylostae | AY960986 | — | 台湾省 |
| Nosema sp.C01 | Pieris rapae | AY383655 | — | 江苏省 |
| Nosema plutellae | Plutella xylostea | AY960987 | — | 台湾省 |
| Nosema spodoptrae | Spodoptera litura | AY747307 | — | 台湾省 |
| Beauveria bassiana | Blattaria | EU334679 | — | 加拿大魁北克省 |
| Nosema antheraeae | Antherea pernyi | — | DQ073396 | 河南省 |
| Nosema disstriae | Malacasoma disstria | — | EU221229 | 加拿大安大略省 |
Table 1 Source of the SSU rDNA and ITS sequences of microsporidia
| 名称 | 宿主 | SSU rDNA GenBank登录号 | ITS GenBank登录号 | 地理来源 |
|---|---|---|---|---|
| YNMZCB1 | Bombyx mori | OQ970549 | OQ974967 | 云南省蒙自市 |
| YNMZCB2 | OQ970550 | OQ974968 | 云南省蒙自市 | |
| YNMZLQ | OQ970551 | OQ974969 | 云南省蒙自市 | |
| YNCXDY | OR597059 | OR597057 | 云南省大姚县 | |
| YNQJLL | OR597056 | OR597058 | 云南省陆良县 | |
| YN1 | JF334596 | JF443647 | 云南省 | |
| YN2 | JF334597 | JF443648 | 云南省 | |
| YN3 | JF334598 | JF443649 | 云南省 | |
| YN4 | JF334599 | JF443650 | 云南省 | |
| SC1 | JF334592 | JF443631 | 四川省 | |
| CQ1 | EU350391 | EU350394 | 重庆市 | |
| GX1 | JF443547 | JF443600 | 广西壮族自治区 | |
| GD1 | JF334582 | JF443619 | 广东省 | |
| AH | GQ334399 | — | 安徽省 | |
| JS | AY616662 | — | 江苏省 | |
| TW | AY209011 | AY209011 | 台湾省 | |
| J-IK | AY259631 | AY259631 | 日本茨城县 | |
| Nosema ceranae | Apis mellifera | DQ486027 | — | 台湾省 |
| Nosema apis | Apis cerana | U97150 | — | 美国加利福尼亚州 |
| Nosema sp.SC | Samia cynthia ricini | FJ767862 | — | 江苏省 |
| Nosema sp.PX1 | Plutella xylostae | AY960986 | — | 台湾省 |
| Nosema sp.C01 | Pieris rapae | AY383655 | — | 江苏省 |
| Nosema plutellae | Plutella xylostea | AY960987 | — | 台湾省 |
| Nosema spodoptrae | Spodoptera litura | AY747307 | — | 台湾省 |
| Beauveria bassiana | Blattaria | EU334679 | — | 加拿大魁北克省 |
| Nosema antheraeae | Antherea pernyi | — | DQ073396 | 河南省 |
| Nosema disstriae | Malacasoma disstria | — | EU221229 | 加拿大安大略省 |
| 分离株 | 核苷酸位点 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 79 | 102 | 294 | 445 | 471 | 558 | 565 | 591 | 640 | 651 | 696 | 703 | 823 | 844 | 861 | |
| YNMZCB1 | G | G | T | G | — | T | G | G | — | C | A | T | G | A | C |
| YNMZCB2 | G | G | C | G | T | T | A | G | — | T | A | C | A | G | T |
| YNMZLQ | A | G | C | G | — | T | A | G | — | T | A | C | G | G | C |
| YNCXDY | G | G | C | C | — | T | A | G | — | T | A | C | G | G | C |
| YNQJLL | G | G | C | G | — | T | A | G | — | T | A | C | G | G | C |
| JS | G | A | C | G | — | G | A | A | C | C | T | C | G | G | C |
Table 2 Differential loci of SSU rDNA gene sequence in N. bombycis
| 分离株 | 核苷酸位点 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 79 | 102 | 294 | 445 | 471 | 558 | 565 | 591 | 640 | 651 | 696 | 703 | 823 | 844 | 861 | |
| YNMZCB1 | G | G | T | G | — | T | G | G | — | C | A | T | G | A | C |
| YNMZCB2 | G | G | C | G | T | T | A | G | — | T | A | C | A | G | T |
| YNMZLQ | A | G | C | G | — | T | A | G | — | T | A | C | G | G | C |
| YNCXDY | G | G | C | C | — | T | A | G | — | T | A | C | G | G | C |
| YNQJLL | G | G | C | G | — | T | A | G | — | T | A | C | G | G | C |
| JS | G | A | C | G | — | G | A | A | C | C | T | C | G | G | C |
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 名称 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.006 | 0.005 | 0.005 | 0.004 | 0.010 | 0.011 | 0.006 | 0.009 | 0.006 | 0.004 | 0.006 | 0.011 | 0.006 | 0.006 | 0.006 | 0.004 | YNMZCB1 | |
| 2 | 99.4 | 0.002 | 0.002 | 0.002 | 0.006 | 0.006 | 0.000 | 0.006 | 0.004 | 0.003 | 0.002 | 0.008 | 0.004 | 0.006 | 0.002 | 0.002 | YNMZCB2 | |
| 3 | 99.5 | 99.8 | 0.002 | 0.001 | 0.007 | 0.007 | 0.002 | 0.006 | 0.003 | 0.001 | 0.002 | 0.007 | 0.003 | 0.005 | 0.002 | 0.001 | YNMZLQ | |
| 4 | 99.5 | 99.8 | 99.8 | 0.001 | 0.007 | 0.007 | 0.002 | 0.006 | 0.003 | 0.002 | 0.002 | 0.007 | 0.003 | 0.005 | 0.002 | 0.001 | YNCXDY | |
| 5 | 99.6 | 99.8 | 99.9 | 99.9 | 0.006 | 0.006 | 0.002 | 0.005 | 0.002 | 0.002 | 0.002 | 0.006 | 0.002 | 0.004 | 0.002 | 0.000 | YNQJLL | |
| 6 | 99.0 | 99.4 | 99.3 | 99.3 | 99.4 | 0.011 | 0.006 | 0.011 | 0.008 | 0.007 | 0.006 | 0.012 | 0.008 | 0.010 | 0.006 | 0.006 | YN1 | |
| 7 | 98.9 | 99.4 | 99.3 | 99.3 | 99.4 | 98.9 | 0.006 | 0.011 | 0.009 | 0.008 | 0.006 | 0.013 | 0.006 | 0.011 | 0.006 | 0.006 | YN2 | |
| 8 | 99.4 | 100.0 | 99.8 | 99.8 | 99.8 | 99.4 | 99.4 | 0.006 | 0.004 | 0.003 | 0.002 | 0.008 | 0.004 | 0.006 | 0.002 | 0.002 | YN3 | |
| 9 | 99.1 | 99.4 | 99.4 | 99.4 | 99.5 | 98.9 | 98.9 | 99.4 | 0.007 | 0.006 | 0.006 | 0.011 | 0.007 | 0.009 | 0.006 | 0.005 | YN4 | |
| 10 | 99.4 | 99.6 | 99.7 | 99.7 | 99.8 | 99.2 | 99.1 | 99.6 | 99.3 | 0.004 | 0.004 | 0.007 | 0.005 | 0.006 | 0.004 | 0.002 | SC1 | |
| 11 | 99.6 | 99.7 | 99.9 | 99.8 | 99.8 | 99.3 | 99.2 | 99.7 | 99.4 | 99.6 | 0.003 | 0.008 | 0.004 | 0.006 | 0.003 | 0.002 | CQ1 | |
| 12 | 99.4 | 99.8 | 99.8 | 99.8 | 99.8 | 99.4 | 99.4 | 99.8 | 99.4 | 99.6 | 99.7 | 0.008 | 0.004 | 0.006 | 0.002 | 0.002 | GX1 | |
| 13 | 98.9 | 99.2 | 99.3 | 99.3 | 99.4 | 98.8 | 98.7 | 99.2 | 98.9 | 99.3 | 99.2 | 99.2 | 0.009 | 0.009 | 0.008 | 0.006 | GD1 | |
| 14 | 99.4 | 99.6 | 99.7 | 99.7 | 99.8 | 99.2 | 99.4 | 99.6 | 99.3 | 99.5 | 99.6 | 99.6 | 99.1 | 0.006 | 0.004 | 0.002 | AH | |
| 15 | 99.4 | 99.4 | 99.5 | 99.5 | 99.6 | 99.0 | 98.9 | 99.4 | 99.1 | 99.4 | 99.4 | 99.4 | 99.1 | 99.4 | 0.006 | 0.004 | JS | |
| 16 | 99.4 | 99.8 | 99.8 | 99.8 | 99.8 | 99.4 | 99.4 | 99.8 | 99.4 | 99.6 | 99.7 | 99.8 | 99.2 | 99.6 | 99.4 | 0.002 | TW | |
| 17 | 99.6 | 99.8 | 99.9 | 99.9 | 100.0 | 99.4 | 99.4 | 99.8 | 99.5 | 99.8 | 99.8 | 99.8 | 99.4 | 99.8 | 99.6 | 99.8 | J-IK |
Table 3 Analysis on sequence homology (bottom left half) and genetic distance (upper right half) of SSU rDNA of N. bombycis from different sericulture regions
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 名称 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.006 | 0.005 | 0.005 | 0.004 | 0.010 | 0.011 | 0.006 | 0.009 | 0.006 | 0.004 | 0.006 | 0.011 | 0.006 | 0.006 | 0.006 | 0.004 | YNMZCB1 | |
| 2 | 99.4 | 0.002 | 0.002 | 0.002 | 0.006 | 0.006 | 0.000 | 0.006 | 0.004 | 0.003 | 0.002 | 0.008 | 0.004 | 0.006 | 0.002 | 0.002 | YNMZCB2 | |
| 3 | 99.5 | 99.8 | 0.002 | 0.001 | 0.007 | 0.007 | 0.002 | 0.006 | 0.003 | 0.001 | 0.002 | 0.007 | 0.003 | 0.005 | 0.002 | 0.001 | YNMZLQ | |
| 4 | 99.5 | 99.8 | 99.8 | 0.001 | 0.007 | 0.007 | 0.002 | 0.006 | 0.003 | 0.002 | 0.002 | 0.007 | 0.003 | 0.005 | 0.002 | 0.001 | YNCXDY | |
| 5 | 99.6 | 99.8 | 99.9 | 99.9 | 0.006 | 0.006 | 0.002 | 0.005 | 0.002 | 0.002 | 0.002 | 0.006 | 0.002 | 0.004 | 0.002 | 0.000 | YNQJLL | |
| 6 | 99.0 | 99.4 | 99.3 | 99.3 | 99.4 | 0.011 | 0.006 | 0.011 | 0.008 | 0.007 | 0.006 | 0.012 | 0.008 | 0.010 | 0.006 | 0.006 | YN1 | |
| 7 | 98.9 | 99.4 | 99.3 | 99.3 | 99.4 | 98.9 | 0.006 | 0.011 | 0.009 | 0.008 | 0.006 | 0.013 | 0.006 | 0.011 | 0.006 | 0.006 | YN2 | |
| 8 | 99.4 | 100.0 | 99.8 | 99.8 | 99.8 | 99.4 | 99.4 | 0.006 | 0.004 | 0.003 | 0.002 | 0.008 | 0.004 | 0.006 | 0.002 | 0.002 | YN3 | |
| 9 | 99.1 | 99.4 | 99.4 | 99.4 | 99.5 | 98.9 | 98.9 | 99.4 | 0.007 | 0.006 | 0.006 | 0.011 | 0.007 | 0.009 | 0.006 | 0.005 | YN4 | |
| 10 | 99.4 | 99.6 | 99.7 | 99.7 | 99.8 | 99.2 | 99.1 | 99.6 | 99.3 | 0.004 | 0.004 | 0.007 | 0.005 | 0.006 | 0.004 | 0.002 | SC1 | |
| 11 | 99.6 | 99.7 | 99.9 | 99.8 | 99.8 | 99.3 | 99.2 | 99.7 | 99.4 | 99.6 | 0.003 | 0.008 | 0.004 | 0.006 | 0.003 | 0.002 | CQ1 | |
| 12 | 99.4 | 99.8 | 99.8 | 99.8 | 99.8 | 99.4 | 99.4 | 99.8 | 99.4 | 99.6 | 99.7 | 0.008 | 0.004 | 0.006 | 0.002 | 0.002 | GX1 | |
| 13 | 98.9 | 99.2 | 99.3 | 99.3 | 99.4 | 98.8 | 98.7 | 99.2 | 98.9 | 99.3 | 99.2 | 99.2 | 0.009 | 0.009 | 0.008 | 0.006 | GD1 | |
| 14 | 99.4 | 99.6 | 99.7 | 99.7 | 99.8 | 99.2 | 99.4 | 99.6 | 99.3 | 99.5 | 99.6 | 99.6 | 99.1 | 0.006 | 0.004 | 0.002 | AH | |
| 15 | 99.4 | 99.4 | 99.5 | 99.5 | 99.6 | 99.0 | 98.9 | 99.4 | 99.1 | 99.4 | 99.4 | 99.4 | 99.1 | 99.4 | 0.006 | 0.004 | JS | |
| 16 | 99.4 | 99.8 | 99.8 | 99.8 | 99.8 | 99.4 | 99.4 | 99.8 | 99.4 | 99.6 | 99.7 | 99.8 | 99.2 | 99.6 | 99.4 | 0.002 | TW | |
| 17 | 99.6 | 99.8 | 99.9 | 99.9 | 100.0 | 99.4 | 99.4 | 99.8 | 99.5 | 99.8 | 99.8 | 99.8 | 99.4 | 99.8 | 99.6 | 99.8 | J-IK |
| 分离株 | 长度/bp | GC含量/% |
|---|---|---|
| YNMZCB1 | 187 | 15.51 |
| YNMZCB2 | 186 | 19.35 |
| YNMZLQ | 180 | 20.56 |
| YNCXDY | 184 | 21.20 |
| YNQJLL | 181 | 18.23 |
| YN1~YN18 | 179~192 | 15.51~21.11 |
| SC1~SC16 | 181~186 | 16.67~18.68 |
| GX1~GX19 | 179~190 | 15.51~21.05 |
| GD1~GD12 | 179~190 | 17.13~20.54 |
| CQ1~CQ10 | 180~187 | 15.51~20.54 |
Table 4 Sequence length and GC content analysis of rDNA-ITS gene sequence of N. bombycis
| 分离株 | 长度/bp | GC含量/% |
|---|---|---|
| YNMZCB1 | 187 | 15.51 |
| YNMZCB2 | 186 | 19.35 |
| YNMZLQ | 180 | 20.56 |
| YNCXDY | 184 | 21.20 |
| YNQJLL | 181 | 18.23 |
| YN1~YN18 | 179~192 | 15.51~21.11 |
| SC1~SC16 | 181~186 | 16.67~18.68 |
| GX1~GX19 | 179~190 | 15.51~21.05 |
| GD1~GD12 | 179~190 | 17.13~20.54 |
| CQ1~CQ10 | 180~187 | 15.51~20.54 |
| 1 | CAPELLA-GUTIÉRREZ S, MARCET-HOUBEN M, GABALDÓN T. Phylogenomics supports microsporidia as the earliest diverging clade of sequenced fungi[J/OL]. BMC Biol., 2012, 10: 47[2024-05-12]. . |
| 2 | HAN B, WEISS L M. Microsporidia: obligate intracellular pathogens within the fungal kingdom[J/OL]. Microbiol. Spectrum., 2017, 5(2): FUNK-0018-2016[2024-05-12]. . |
| 3 | STENTIFORD G D, FEIST S W, STONE D M, et al.. Microsporidia: diverse, dynamic, and emergent pathogens in aquatic systems[J]. Trends Parasitol., 2013, 29(11): 567-578. |
| 4 | 刘吉平,曾玲.微孢子虫生物多样性研究的述评[J].昆虫知识,2006,43(2):153-158. |
| LIU J P, ZENG L. An overview of research on the microsporidian biodiversity[J]. Chin. Bull. Entomol., 2006, 43(2): 153-158. | |
| 5 | CALI A, WEISS L M, TAKVORIAN P M. An analysis of the microsporidian genus Brachiola, with comparisons of human and insect isolates of Brachiola algerae[J]. J. Eukaryot. Microbiol., 2004, 51(6): 678-685. |
| 6 | WEISS L M. The first united workshop on microsporidia from invertebrate and vertebrate hosts[J]. Folia Parasitol. Praha., 2005, 52(1-2): 1-7. |
| 7 | BAKER M D, VOSSBRINCK C R, DIDIER E S, et al.. Small subunit ribosomal DNA phylogeny of various microsporidia with emphasis on AIDS related forms[J]. J. Eukaryot. Microbiol., 1995, 42(5): 564-570. |
| 8 | HIRT R P, LOGSDON J M, HEALY B, et al.. Microsporidia are related to Fungi: evidence from the largest subunit of RNA polymerase Ⅱ and other proteins[J]. Proc. Natl. Acad. Sci. USA, 1999, 96(2): 580-585. |
| 9 | CORNMAN R S, CHEN Y P, SCHATZ M C, et al.. Genomic analyses of the microsporidian Nosema ceranae, an emergent pathogen of honey bees[J/OL]. PLoS Pathog., 2009, 5(6): e1000466[2024-05-12]. . |
| 10 | PAN G, XU J, LI T, et al.. Comparative genomics of parasitic silkworm microsporidia reveal an association between genome expansion and host adaptation[J/OL]. BMC Genom., 2013, 14: 186[2024-05-12]. . |
| 11 | XU J, HE Q, MA Z, et al.. The genome of Nosema sp. isolate YNPr: a comparative analysis of genome evolution within the Nosema/Vairimorpha clade [J/OL]. PLoS One, 2016, 11(9): e0162336[2024-05-12]. . |
| 12 | BALCH W E, MAGRUM L J, FOX G E, et al.. An ancient divergence among the bacteria[J]. J. Mol. Evol., 1977, 9(4): 305-311. |
| 13 | KEL C E N, ISSI I V. Effect of changes in insect hosts on the pathogenicity and spore formation in microsporidia[J]. Parazitologigya, 1991, 25(6): 512-519. |
| 14 | 黄少康,鲁兴萌.家蚕微粒子虫(Nosema bombycis)与其形态变异株的侵染性及孢子表面蛋白的比较研究[J].中国农业科学,2004,37(11):1682-1687. |
| HUANG S K, LU X M. Comparative study on the infectivity and spore surface protein of Nosema bombycis and its morphological variant strain[J]. Sci. Agric. Sin., 2004, 37(11): 1682-1687. | |
| 15 | 张素贞,何超,王艳丽,等.重庆地区蜜蜂微孢子虫的鉴定及分子遗传多样性分析[J].西南农业学报,2015,28(5):2323-2330. |
| ZHANG S Z, HE C, WANG Y L, et al.. Identification and genetic diversity of microsporidia Nosema ceranae isolated from Chongqing based on multilocus molecular markers[J]. Southwest China J. Agric. Sci., 2015, 28(5): 2323-2330. | |
| 16 | HATAKEYAMA Y, ODA H, TSUNODA R, et al.. Genome profiling implies high genetic diversity in microsporidia isolated from the common cutworm, Spodoptera litura (Lepidoptera: Noctuidae), in Vietnam[J]. Appl. Entomol. Zool., 2011, 46(3): 293-299. |
| 17 | XING D, LI Q, ZHANG J, et al.. Phylogenetic analysis of the complete rRNA gene sequence of Nosema sp. SE isolated from the beet armyworm Spodoptera exigua [J]. J. Parasitol., 2019, 105(6): 878-881. |
| 18 | 时连辉,邱宝利,高绘菊.不同地域来源的家蚕微孢子虫DNA特异片段序列及同源性分析[J].蚕业科学,2003,29(4):384-386. |
| SHI L H, QIU B L, GAO H J. Study on the comparision of sequence similarity of three Nosema bombycis geographical strains[J]. Acta Sericologica Sin., 2003, 29(4): 384-386. | |
| 19 | 申子刚,潘国庆,许金山,等.重庆地区家蚕微孢子虫遗传多态性分析[J].自然科学进展,2008,18(5):579-586. |
| 20 | 黄旭华,罗梅兰,汤庆坤,等.家蚕病原性微孢子虫多样性调查分析[J].南方农业学报,2018,49(6):1208-1214. |
| HUANG X H, LUO M L, TANG Q K, et al.. Diversity of pathogenic microsporidan of Bombyx mori [J]. J. South. Agric., 2018, 49(6): 1208-1214. | |
| 21 | LIU H, PAN G, LUO B, et al.. Intraspecific polymorphism of rDNA among five Nosema bombycis isolates from different geographic regions in China[J]. J. Invertebr. Pathol., 2013, 113(1): 63-69. |
| 22 | 周成,潘国庆,万永继,等.家蚕微孢子虫N. bombycis分离纯化方法的优化[J].蚕学通讯,2002,22(1):7-9+6. |
| ZHOU C, PAN G Q, WAN Y J, et al.. Optimization of the procedures of isolation and purification of nosema bombycis in silkworm[J]. Newsl. Sericultural. Sci., 2002, 22(1): 7-9+6. | |
| 23 | 潘敏慧, 万永继, 鲁成. 不同种类微孢子虫DNA制备方法的研究[J]. 西南农业大学学报, 2001, 23(2):111-112. |
| 24 | HUANG W F, BOCQUET M, LEE K C, et al.. The comparison of rDNA spacer regions of Nosema ceranae isolates from different hosts and locations[J]. J. Invertebr. Pathol., 2008, 97(1): 9-13. |
| 25 | KUMAR S, STECHER G, TAMURA K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets[J]. Mol. Biol. Evol., 2016, 33(7): 1870-1874. |
| 26 | HUANG W F, TSAI S J, LO C F, et al.. The novel organization and complete sequence of the ribosomal RNA gene of Nosema bombycis [J]. Fungal. Genet. Biol., 2004, 41(5): 473-481. |
| 27 | ZHANG Y H, TANG F F, SHAO Y L, et al.. Molecular epidemiology of Bombyx mori nucleopolyhedrovirus in Yunnan sericulture region, China[J]. Sci. Asia, 2019, 45(4):332-341. |
| 28 | 王敏,许金山,王林玲,等.两株不同地域的家蚕微孢子虫分离株的细胞侵染力比较及遗传多样性分析[J].遗传,2009,31(11):1121-1126. |
| WANG M, XU J S, WANG L L, et al.. Pathogenicity and genetic divergence of two isolates of microsporidia Nosema bombycis [J]. Hereditas, 2009, 31(11): 1121-1126. | |
| 29 | 郑祥明,杨琼,方定坚,等.广东省昆虫微孢子虫资源调查及交叉感染的研究[J].蚕业科学,2003,29(4):380-383. |
| ZHENG X M, YANG Q, FANG D J, et al.. Studies on insect microsporidia resources in Guangdong Province and cross infection among some insects[J]. Acta Sericol. Sin., 2003, 29(4): 380-383. | |
| 30 | 黄旭华,潘志新,朱方容,等.广西野外昆虫微孢子虫对家蚕交叉感染情况调查[J].广西蚕业,2010,47(4):15-20. |
| HUANG X H, PAN Z X, ZHU F R, et al.. Investigation on cross-infection of insect microsporidia to silkworm in Guangxi[J]. Guangxi Seric., 2010, 47(4): 15-20. | |
| 31 | 陈世良,肖圣燕,潘秋玲,等.梨花迁粉蝶微孢子虫对家蚕的侵染力与胚传性[J].昆虫学报,2017,60(2):155-162. |
| CHEN S L, XIAO S Y, PAN Q L, et al.. Infectivity and transmissibility of Nosema sp. CP isolated from Catopsilia pyranthe (Lepidoptera: Pieridae) in the domestic silkworm (Bombyx mori)[J]. Acta Entomol. Sin., 2017, 60(2): 155-162. |
| [1] | Xiaorui YAN, Guozhen XUE, Xiaoxia YANG, Yu WANG, Chenhui DU, Shuosheng ZHANG, Jiquan LIU. ISSR Genetic Diversity Analysis of Bupleurum chinense from Different Populations [J]. Current Biotechnology, 2023, 13(6): 919-924. |
| [2] | Qinxue GAO, Wenke YANG, Yonggang MENG, Nashier YASHENGJIANG, Kuwaxi MAIMAITITUERGAN. Genetic Diversity of Xinjiang Kizilsu Kirghiz Region Pamir Yaks Based on Mitochondrial COⅠ Gene [J]. Current Biotechnology, 2022, 12(4): 568-576. |
| [3] | LI Jing1, WANG Li1, DAO Min2, HU Ping1, XIAO Yi1, HAN Jianlin1, WANG Yutao1*. Genetic Diversity and Phylogenetic Analysis on mtDNA Cytb in Yak of Karakorum-Pamir Region [J]. Curr. Biotech., 2019, 9(5): 509-517. |
| [4] | ZHANG Zhi-ming1, TANG Cai-guo2,3, YANG San-wei1, QIAO Lin-yi1, CHANG Jian-zhong1, ZHAO Cui-rong4,5*, ZHENG Jun1*. Advances on Gene Discovery and Utilization of Wild Relatives of Triticum aestivum L. [J]. Curr. Biotech., 2016, 6(5): 305-311. |
| [5] | DONG Zhi-gang1§, LIU Zheng-hai2§, LI Xiao-mei1, . Application of SSR Markers in Variety Identification and Genetic Breeding of Vitis vinifera [J]. Curr. Biotech., 2016, 6(2): 137-140. |
| [6] | HE Zhen1, YUN Xiao-dong1, WU Kai1, JI Hu-tai2, CHANG Jian-zhong1, QIAO Lin-yi1*, ZHENG Jun2*. Progress on Germplasm Resources Genetic Diversity of Soybean [J]. Curr. Biotech., 2015, 5(2): 103-108. |
| [7] | WANG Shi-feng, LI Jing, WANG Yu-tao. Genetic Diversity and Phylogeny Analysis of Duolang Sheep Based on Mitochondrial DNA-loop Variation [J]. Curr. Biotech., 2014, 4(6): 429-434. |
| [8] | HE Hu-yi, TAN Guan-ning, HE Xin-min*, HE Hai-wang, LI Li-shu, TANG Zhou-ping, WANG Hui. Application of Several PCR-based Molecular Markers in Sweet Potato Genetic Diversity Researches [J]. Curr. Biotech., 2014, 4(4): 245-250. |
| [9] | WANG Han-yu1, Du Yan-wei2, LI Ping2, ZHANG Xi-wen2, ZHU Jing-ying1, ZHAO Jin-feng2, YU Ai-li1*. Genetic Relationship of 40 Maize Inbred Lines Based on ISSR [J]. journal1, 2011, 1(3): 214-218. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||