Current Biotechnology ›› 2024, Vol. 14 ›› Issue (4): 618-630.DOI: 10.19586/j.2095-2341.2024.0024
• Articles • Previous Articles Next Articles
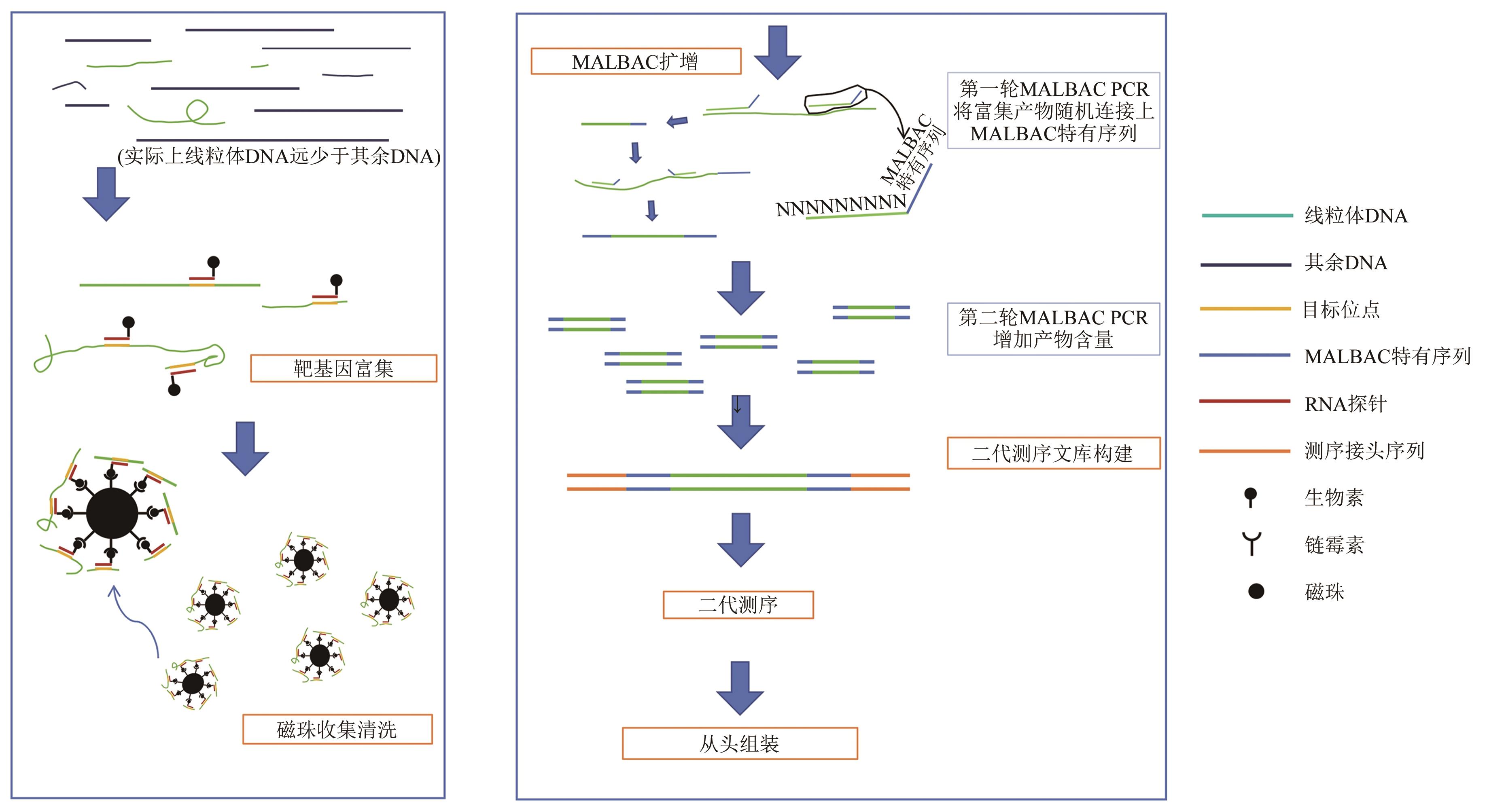
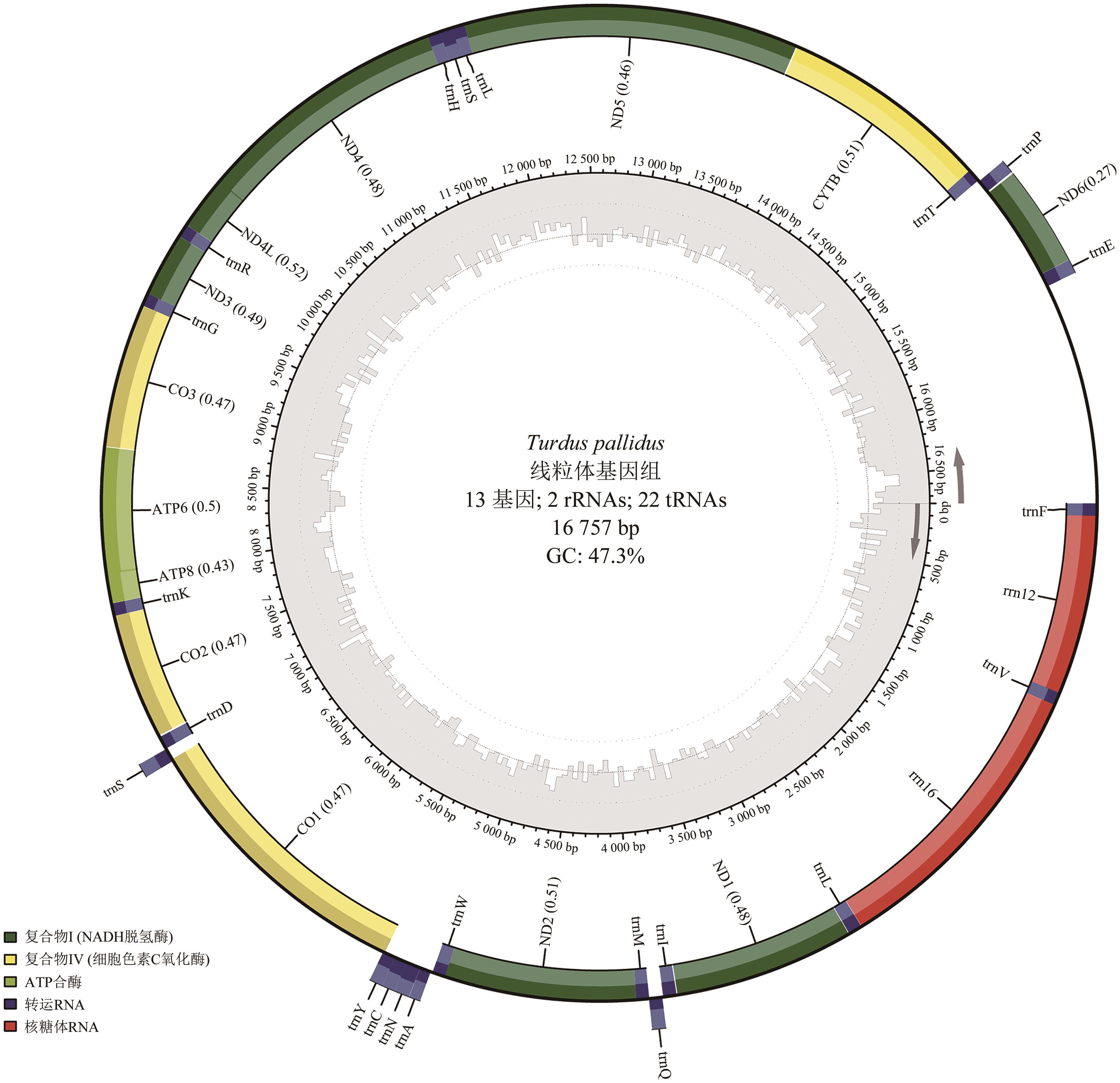
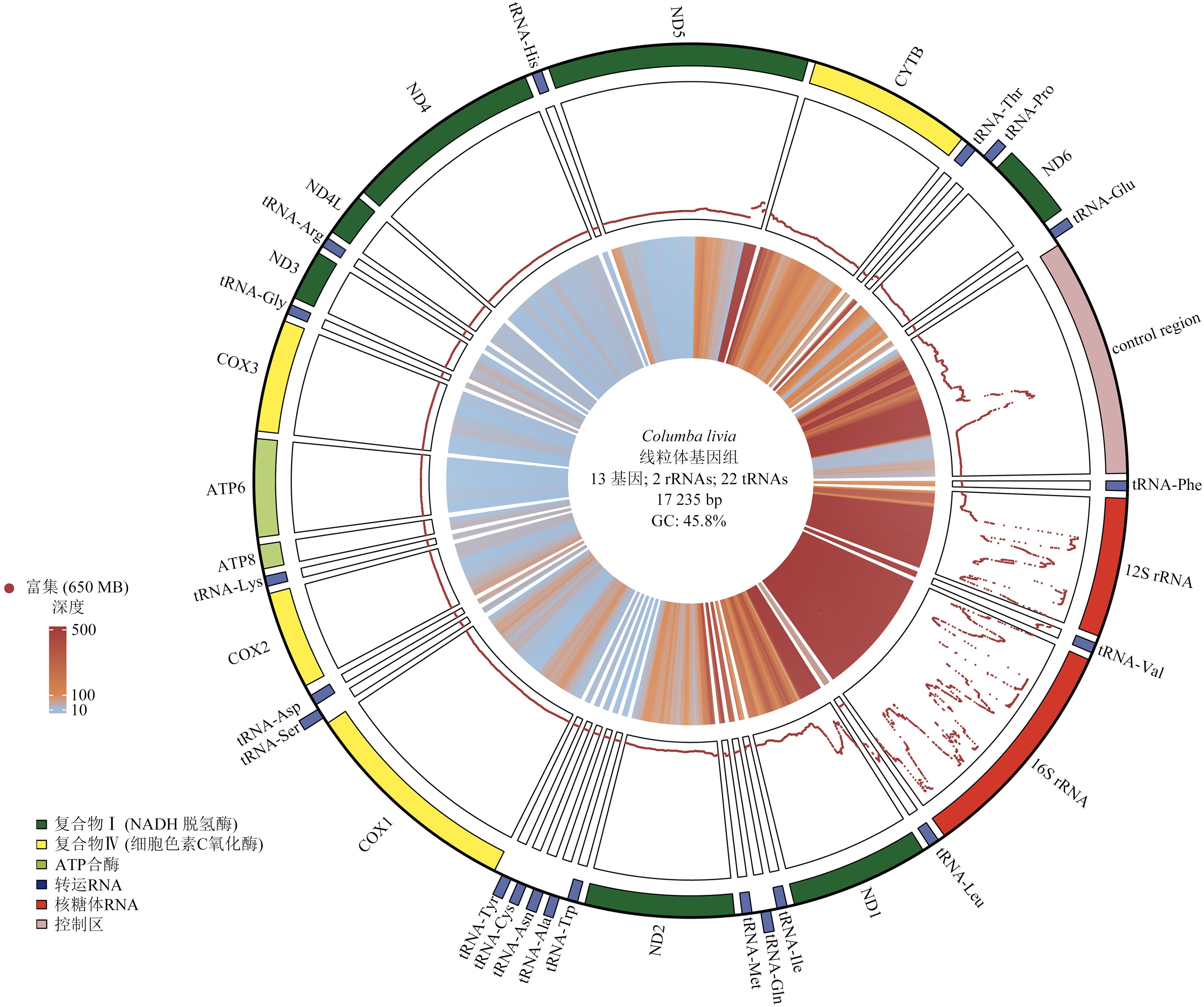
Rapid Acquisition of Avian Mitochondrial Genome Based on Gene Capture Method
Bingying QIU( ), Xueyao CHEN, Hui WANG, Chenhong LI, Dongsheng ZHANG(
), Xueyao CHEN, Hui WANG, Chenhong LI, Dongsheng ZHANG( )
)
- College of Fisheries and Life Science,Shanghai Ocean University,Shanghai 201306,China
-
Received:2024-02-20Accepted:2024-03-05Online:2024-07-25Published:2024-08-07 -
Contact:Dongsheng ZHANG
基于基因富集的鸟类线粒体基因组快速获取方法研究
- 上海海洋大学水产与生命学院,上海 201306
-
通讯作者:张东升 -
作者简介:邱冰滢 E-mail:m210100084@st.shou.edu.cn; -
基金资助:国家重点研发计划项目(2022YFC2601301)
CLC Number:
Cite this article
Bingying QIU, Xueyao CHEN, Hui WANG, Chenhong LI, Dongsheng ZHANG. Rapid Acquisition of Avian Mitochondrial Genome Based on Gene Capture Method[J]. Current Biotechnology, 2024, 14(4): 618-630.
邱冰滢, 陈雪瑶, 王辉, 李晨虹, 张东升. 基于基因富集的鸟类线粒体基因组快速获取方法研究[J]. 生物技术进展, 2024, 14(4): 618-630.
share this article
| 序号 | 样本编号 | 组织 |
|---|---|---|
| 1 | P11-1 | 趾部骨骼和皮肤 |
| 2 | P14-1 | 趾部骨骼和皮肤 |
| 3 | P4-1 | 肌肉 |
| 4 | P10-1 | 趾部骨骼和皮肤 |
| 5 | P8-1 | 趾部骨骼和皮肤 |
| 6 | P9-1 | 趾部骨骼和皮肤 |
| 7 | P1-1 | 肌肉 |
| 8 | P3-1 | 肌肉 |
| 9 | P12-1 | 肌肉 |
| 10 | P12-3 | 肌肉 |
| 11 | P6-1 | 肌肉 |
| 12 | P7-1 | 肌肉 |
| 13 | P13-1 | 肌肉 |
| 14 | P15-1 | 肌肉 |
| 15 | N10-1 | 肌肉 |
| 16 | GS-8 | 肌肉 |
| 17 | QP-2 | 趾部骨骼和皮肤 |
| 18 | QP-4 | 趾部骨骼和皮肤 |
| 19 | N2-1 | 趾部骨骼和皮肤 |
Table 1 The list of bird samples in this study
| 序号 | 样本编号 | 组织 |
|---|---|---|
| 1 | P11-1 | 趾部骨骼和皮肤 |
| 2 | P14-1 | 趾部骨骼和皮肤 |
| 3 | P4-1 | 肌肉 |
| 4 | P10-1 | 趾部骨骼和皮肤 |
| 5 | P8-1 | 趾部骨骼和皮肤 |
| 6 | P9-1 | 趾部骨骼和皮肤 |
| 7 | P1-1 | 肌肉 |
| 8 | P3-1 | 肌肉 |
| 9 | P12-1 | 肌肉 |
| 10 | P12-3 | 肌肉 |
| 11 | P6-1 | 肌肉 |
| 12 | P7-1 | 肌肉 |
| 13 | P13-1 | 肌肉 |
| 14 | P15-1 | 肌肉 |
| 15 | N10-1 | 肌肉 |
| 16 | GS-8 | 肌肉 |
| 17 | QP-2 | 趾部骨骼和皮肤 |
| 18 | QP-4 | 趾部骨骼和皮肤 |
| 19 | N2-1 | 趾部骨骼和皮肤 |
| 序号 | 中文名 | 拉丁名 | 目 | 科 | GenBank登陆号 |
|---|---|---|---|---|---|
| 1 | 珠颈斑鸠 | Spilopelia chinensis | 鸽形目 | 鸠鸽科 | NC_026459.1 |
| 2 | 黑水鸡 | Gallinula chloropus | 鹤形目 | 秧鸡科 | NC_015236.1 |
| 3 | 鸡 | Gallus gallus | 鸡形目 | 雉科 | NC_053523.1 |
| 4 | 普通鸬鹚 | Phalacrocorax carbo | 鲣鸟目 | 鸬鹚科 | NC_027267.1 |
| 5 | 大斑啄木鸟 | Dendrocopos major | 䴕形目 | 啄木鸟科 | NC_028174.1 |
| 6 | 小䴙䴘 | Tachybaptus ruficollis | 䴙䴘目 | 䴙䴘科 | NC_024594.1 |
| 7 | 白鹭 | Egretta garzetta | 鹈形目 | 鹭科 | NC_023981.1 |
| 8 | 戴胜 | Upupa epops | 犀鸟目 | 戴胜科 | NC_028178.1 |
| 9 | 白头鹎 | Pycnonotus sinensis | 雀形目 | 鹎科 | NC_013838.1 |
| 10 | 麻雀 | Passer montanus | 雀形目 | 雀科 | NC_024821.1 |
| 11 | 喜鹊 | Pica pica | 雀形目 | 鸦科 | NC_015200.1 |
| 12 | 白鹡鸰 | Motacilla alba | 雀形目 | 鹡鸰科 | NC_029229.1 |
| 13 | 乌鸫 | Turdus merula | 雀形目 | 鸫科 | NC_028188.1 |
| 14 | 棕背伯劳 | Lanius schach | 雀形目 | 伯劳科 | NC_030604.1 |
| 15 | 家燕 | Hirundo rustica | 雀形目 | 燕科 | NC_050297.1 |
| 16 | 鹊鸲 | Copsychus saularis | 雀形目 | 鹟科 | NC_030603.1 |
| 17 | 八哥 | Acridotheres cristatellus | 雀形目 | 椋鸟科 | NC_015613.1 |
| 18 | 斑胸草雀 | Taeniopygia guttata | 雀形目 | 梅花雀科 | NC_007897.1 |
Table 2 The species and GenBank access ID of the reference mitochondrial genome
| 序号 | 中文名 | 拉丁名 | 目 | 科 | GenBank登陆号 |
|---|---|---|---|---|---|
| 1 | 珠颈斑鸠 | Spilopelia chinensis | 鸽形目 | 鸠鸽科 | NC_026459.1 |
| 2 | 黑水鸡 | Gallinula chloropus | 鹤形目 | 秧鸡科 | NC_015236.1 |
| 3 | 鸡 | Gallus gallus | 鸡形目 | 雉科 | NC_053523.1 |
| 4 | 普通鸬鹚 | Phalacrocorax carbo | 鲣鸟目 | 鸬鹚科 | NC_027267.1 |
| 5 | 大斑啄木鸟 | Dendrocopos major | 䴕形目 | 啄木鸟科 | NC_028174.1 |
| 6 | 小䴙䴘 | Tachybaptus ruficollis | 䴙䴘目 | 䴙䴘科 | NC_024594.1 |
| 7 | 白鹭 | Egretta garzetta | 鹈形目 | 鹭科 | NC_023981.1 |
| 8 | 戴胜 | Upupa epops | 犀鸟目 | 戴胜科 | NC_028178.1 |
| 9 | 白头鹎 | Pycnonotus sinensis | 雀形目 | 鹎科 | NC_013838.1 |
| 10 | 麻雀 | Passer montanus | 雀形目 | 雀科 | NC_024821.1 |
| 11 | 喜鹊 | Pica pica | 雀形目 | 鸦科 | NC_015200.1 |
| 12 | 白鹡鸰 | Motacilla alba | 雀形目 | 鹡鸰科 | NC_029229.1 |
| 13 | 乌鸫 | Turdus merula | 雀形目 | 鸫科 | NC_028188.1 |
| 14 | 棕背伯劳 | Lanius schach | 雀形目 | 伯劳科 | NC_030604.1 |
| 15 | 家燕 | Hirundo rustica | 雀形目 | 燕科 | NC_050297.1 |
| 16 | 鹊鸲 | Copsychus saularis | 雀形目 | 鹟科 | NC_030603.1 |
| 17 | 八哥 | Acridotheres cristatellus | 雀形目 | 椋鸟科 | NC_015613.1 |
| 18 | 斑胸草雀 | Taeniopygia guttata | 雀形目 | 梅花雀科 | NC_007897.1 |
| 序号 | 样本编号 | 物种 | 学名 | GenBank检索编号 |
|---|---|---|---|---|
| 1 | P11-1 | 黄腹柳莺 | Phylloscopus affinis | OR757045 |
| 2 | P14-1 | 棕头鸦雀 | Sinosuthora webbiana | OR757048 |
| 3 | P4-1 | 白眉鹀 | Emberiza tristrami | OR750852 |
| 4 | P10-1 | 黄喉鹀 | Emberiza elegans | OR493959 |
| 5 | P8-1 | 红胁蓝尾鸲 | Tarsiger cyanurus | OR493959 |
| 6 | P9-1 | 白腹蓝鹟 | Cyanoptila cyanomelana | OR757044 |
| 7 | P1-1 | 虎斑地鸫 | Zoothera aurea | OR757050 |
| 8 | P3-1 | 乌鸫 | Turdus mandarinus | OR493960 |
| 9 | P12-1 | 白腹鸫 | Turdus pallidus | OR493961 |
| 10 | P12-3 | 白腹鸫 | Turdus pallidus | OR493962 |
| 11 | P6-1 | 灰背鸫 | Turdus hortulorum | OR493963 |
| 12 | P7-1 | 乌灰鸫 | Turdus cardis | OR757043 |
| 13 | P13-1 | 斑鸫 | Turdus eunomus | OR757047 |
| 14 | P15-1 | 白头鹎 | Pycnonotus sinensis | OR757053 |
| 15 | N10-1 | 黄苇鳽 | Ixobrychus sinensis | OR757051 |
| 16 | GS-8 | 白鹭 | Egretta garzetta | OR757052 |
| 17 | QP-2 | 牛背鹭 | Bubulcus ibis | OR757054 |
| 18 | QP-4 | 牛背鹭 | Bubulcus ibis | OR757055 |
| 19 | N2-1 | 鸽 | Columba livia | OR757049 |
Table 3 The mitochondrial genomes of 19 bird samples corresponded to GenBank access ID
| 序号 | 样本编号 | 物种 | 学名 | GenBank检索编号 |
|---|---|---|---|---|
| 1 | P11-1 | 黄腹柳莺 | Phylloscopus affinis | OR757045 |
| 2 | P14-1 | 棕头鸦雀 | Sinosuthora webbiana | OR757048 |
| 3 | P4-1 | 白眉鹀 | Emberiza tristrami | OR750852 |
| 4 | P10-1 | 黄喉鹀 | Emberiza elegans | OR493959 |
| 5 | P8-1 | 红胁蓝尾鸲 | Tarsiger cyanurus | OR493959 |
| 6 | P9-1 | 白腹蓝鹟 | Cyanoptila cyanomelana | OR757044 |
| 7 | P1-1 | 虎斑地鸫 | Zoothera aurea | OR757050 |
| 8 | P3-1 | 乌鸫 | Turdus mandarinus | OR493960 |
| 9 | P12-1 | 白腹鸫 | Turdus pallidus | OR493961 |
| 10 | P12-3 | 白腹鸫 | Turdus pallidus | OR493962 |
| 11 | P6-1 | 灰背鸫 | Turdus hortulorum | OR493963 |
| 12 | P7-1 | 乌灰鸫 | Turdus cardis | OR757043 |
| 13 | P13-1 | 斑鸫 | Turdus eunomus | OR757047 |
| 14 | P15-1 | 白头鹎 | Pycnonotus sinensis | OR757053 |
| 15 | N10-1 | 黄苇鳽 | Ixobrychus sinensis | OR757051 |
| 16 | GS-8 | 白鹭 | Egretta garzetta | OR757052 |
| 17 | QP-2 | 牛背鹭 | Bubulcus ibis | OR757054 |
| 18 | QP-4 | 牛背鹭 | Bubulcus ibis | OR757055 |
| 19 | N2-1 | 鸽 | Columba livia | OR757049 |
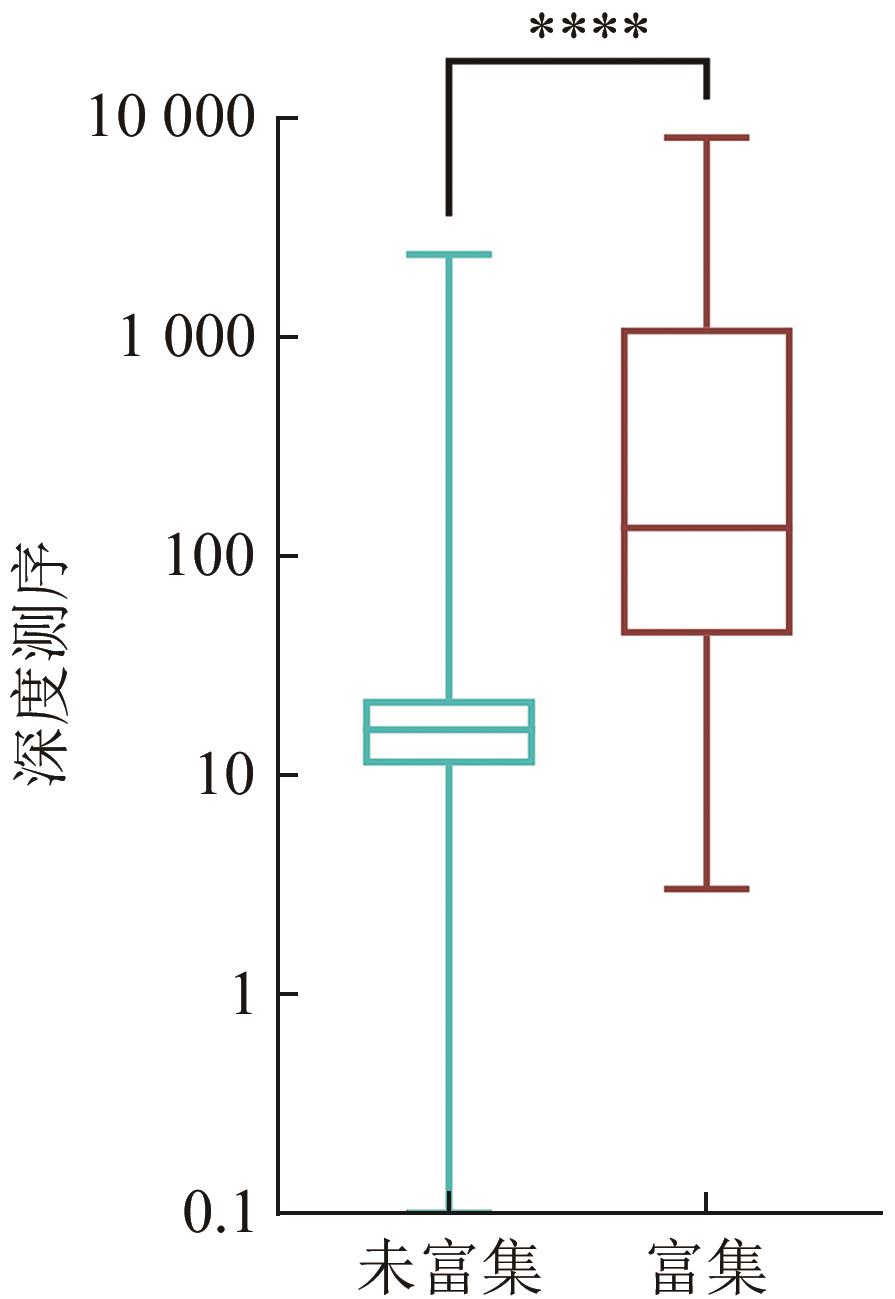
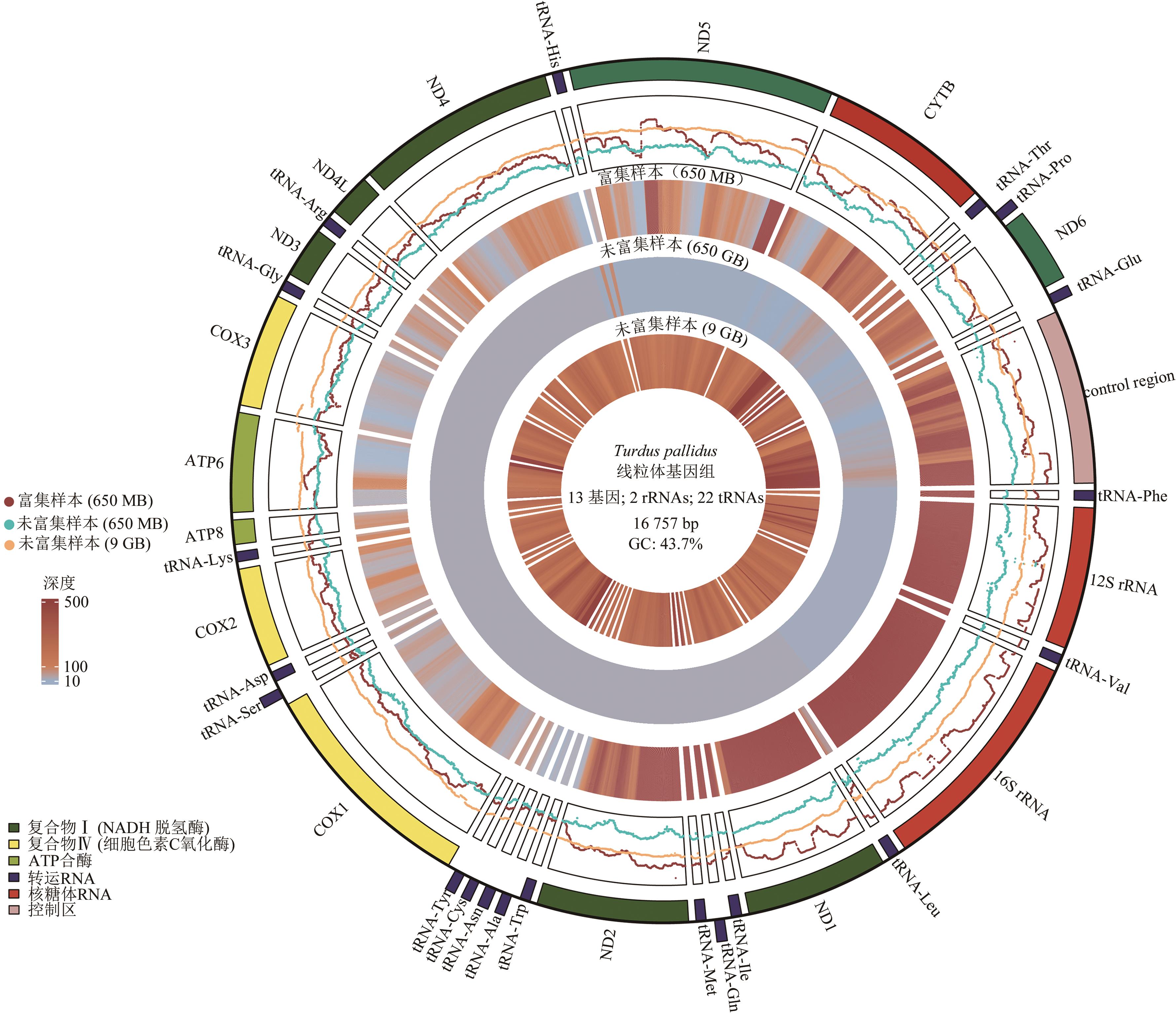
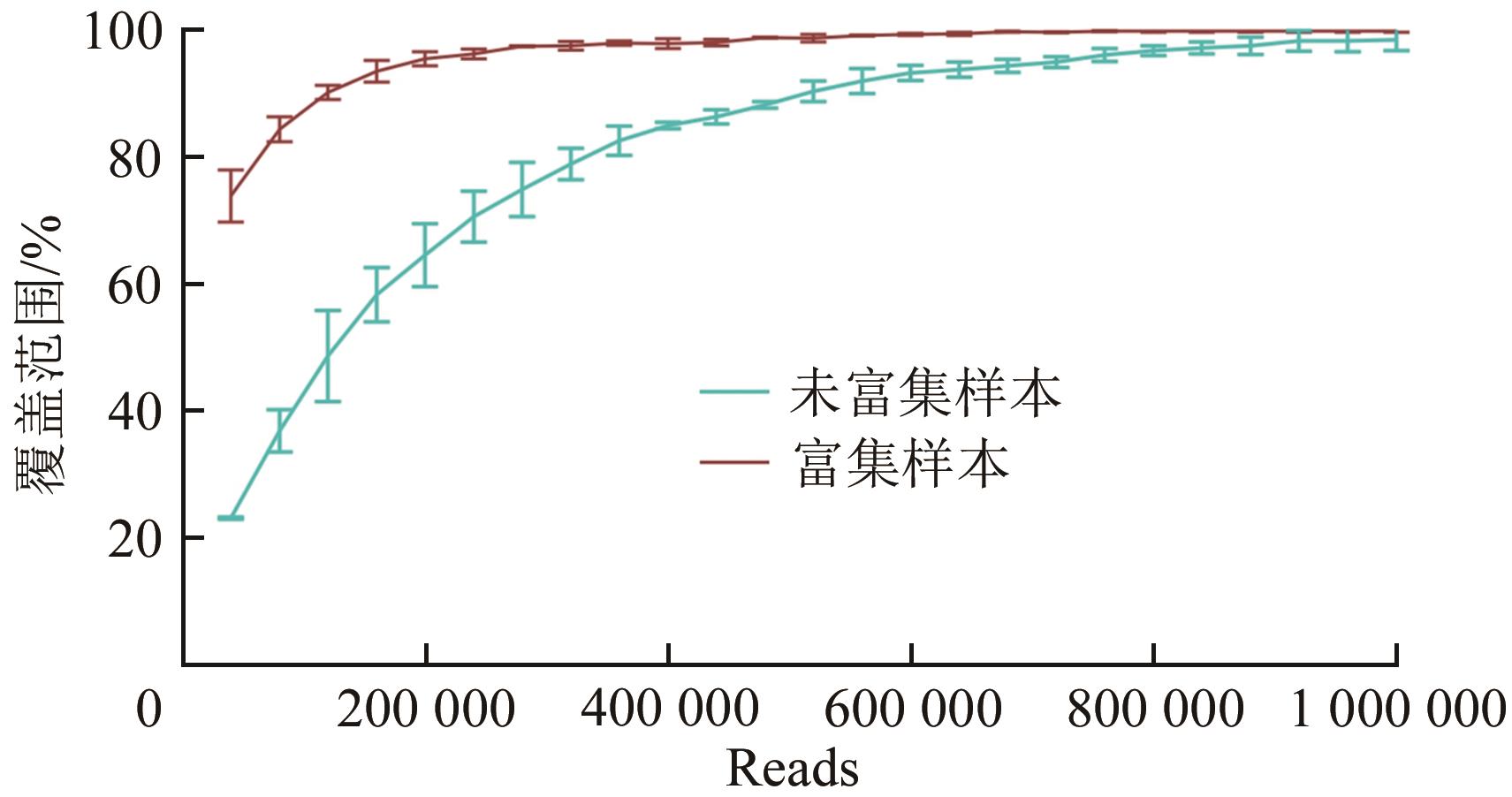
| 参数 | 不富集,直接建库 | 先富集,后建库 | |
|---|---|---|---|
| 样本编号 | P12-1 | ||
| 物种 | 白腹鸫(Turdus pallidus) | ||
| 线粒体基因组长度/bp | 16 757 | ||
| 读序数(Reads数) | 62 270 514 | 4 330 016 | |
| 测序量 | 约9 GB | 650 MB | |
| 比对上的读序数 | 67 954 | 4 776 | 636 222 |
| 比对上的读序数占比% | 0.11% | 0.11% | 14.69% |
| 最高深度 | 8 000 | 2 345 | 8 009 |
| 最低深度 | 5 | 0 | 3 |
| 平均深度 | 269.47 | 21.20 | 1 316.84 |
| 深度为0的碱基数 | 0 | 74 | 0 |
| 覆盖度% | 100% | 99.56% | 100% |
| 富集倍率 | — | 1 | 62.38 |
Table 4 Cover degree and deepth of the two methods
| 参数 | 不富集,直接建库 | 先富集,后建库 | |
|---|---|---|---|
| 样本编号 | P12-1 | ||
| 物种 | 白腹鸫(Turdus pallidus) | ||
| 线粒体基因组长度/bp | 16 757 | ||
| 读序数(Reads数) | 62 270 514 | 4 330 016 | |
| 测序量 | 约9 GB | 650 MB | |
| 比对上的读序数 | 67 954 | 4 776 | 636 222 |
| 比对上的读序数占比% | 0.11% | 0.11% | 14.69% |
| 最高深度 | 8 000 | 2 345 | 8 009 |
| 最低深度 | 5 | 0 | 3 |
| 平均深度 | 269.47 | 21.20 | 1 316.84 |
| 深度为0的碱基数 | 0 | 74 | 0 |
| 覆盖度% | 100% | 99.56% | 100% |
| 富集倍率 | — | 1 | 62.38 |
| 方法 | 深度 | Z | P |
|---|---|---|---|
| 未富集 | 16(11~22) | -140.10 | 0 |
| 富集 | 133(43~1085) |
Table 5 Mann-Whitney U test results of capture and non-capture samples
| 方法 | 深度 | Z | P |
|---|---|---|---|
| 未富集 | 16(11~22) | -140.10 | 0 |
| 富集 | 133(43~1085) |
| 1 | JIA Y, QIU G, CAO C, et al.. Mitochondrial genome and phylogenetic analysis of Chaohu duck[J/OL]. Gene, 2023, 851: 147018[2024-05-03]. . |
| 2 | CHEN X, NI G, HE K, et al.. Capture hybridization of long-range DNA fragments for high-throughput sequencing[J]. Meth. Mol. Biol., 2018, 1754: 29-44. |
| 3 | GREGORY R, NOBLE D, FIELD R, et al.. Using birds as indicators of biodiversity[J]. Ornis Hung., 2003, 12: 11-24. |
| 4 | LI C, HOFREITER M, STRAUBE N, et al.. Capturing protein-coding genes across highly divergent species[J]. Biotechniques, 2013, 54(6): 321-326. |
| 5 | 徐茹晖,张卫平,张东升.基于公民科学方式的上海南汇东滩湿地鸟类多样性研究[J].湿地科学与管理,2022,18(3):16-20. |
| XU R H, ZHANG W P, ZHANG D S. A baseline inventory on bird diversity of Nanhui Dongtan wetland in Shanghai using citizen scientists[J]. Wetl. Sci. Manag., 2022, 18(3): 16-20. | |
| 6 | TAMURA K, STECHER G, KUMAR S. MEGA11: molecular evolutionary genetics analysis version 11[J]. Mol. Biol. Evol., 2021, 38(7): 3022-3027. |
| 7 | 陈文隽. 获得脊椎动物线粒体基因组新方法的开发与改进[D]. 上海:上海海洋大学, 2022. |
| 8 | MASON V C, LI G, HELGEN K M, et al.. Efficient cross-species capture hybridization and next-generation sequencing of mitochondrial genomes from noninvasively sampled museum specimens[J]. Genome Res., 2011, 21(10): 1695-1704. |
| 9 | ZONG C, LU S, CHAPMAN A R, et al.. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell[J]. Science, 2012, 338(6114): 1622-1626. |
| 10 | ROHLAND N, REICH D. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture[J]. Genome Res., 2012, 22(5): 939-946. |
| 11 | JIN J J, YU W B, YANG J B, et al.. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes[J/OL]. Genome Biol., 2020, 21(1): 241[2024-05-03]. . |
| 12 | CAMACHO C, COULOURIS G, AVAGYAN V, et al.. BLAST+: architecture and applications[J/OL]. BMC Bioinform., 2009, 10: 421[2024-05-03]. . |
| 13 | DIERCKXSENS N, MARDULYN P, SMITS G. NOVOPlasty: de novo assembly of organelle genomes from whole genome data[J/OL]. Nucleic Acids Res., 2017, 45(4): e18[2024-05-03]. . |
| 14 | BERNT M, DONATH A, JÜHLING F, et al.. MITOS: improved de novo metazoan mitochondrial genome annotation[J]. Mol. Phylogenet. Evol., 2013, 69(2): 313-319. |
| 15 | ALLIO R, SCHOMAKER-BASTOS A, ROMIGUIER J, et al.. MitoFinder: efficient automated large-scale extraction of mitogenomic data in target enrichment phylogenomics[J]. Mol. Ecol. Resour., 2020, 20(4): 892-905. |
| 16 | LI H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM[J]. arXiv, 2013: 1303.3997. |
| 17 | LI H, HANDSAKER B, WYSOKER A, et al. The Sequence Alignment/Map format and SAMtools[J]. Bioinformatics, 2009, 25(16): 2078-2079. |
| 18 | SHEN W, LE S, LI Y, et al.. SeqKit: a cross-platform and ultrafast toolkit for FASTA/Q file manipulation[J/OL]. PLoS One, 2016, 11(10): e0163962[2024-05-03]. . |
| 19 | LEHWARK P, GREINER S. GB2sequin - A file converter preparing custom GenBank files for database submission[J]. Genomics, 2019, 111(4): 759-761. |
| 20 | 纪艺,徐晓丽,姜媛媛,等.基于数字PCR的不同品种鸭组织中线粒体与核DNA拷贝数差异研究[J].生物技术通报,2020,36(5):86-91. |
| JI Y, XU X L, JIANG Y Y, et al.. Copy number variations of mitochondrial DNA and genomic DNA from different tissues of duck based on digital PCR[J]. Biotechnol. Bull., 2020, 36(5): 86-91. | |
| 21 | SORENSON M D, AST J C, DIMCHEFF D E, et al.. Primers for a PCR-based approach to mitochondrial genome sequencing in birds and other vertebrates[J]. Mol. Phylogenet. Evol., 1999, 12(2): 105-114. |
| 22 | BRISCOE A G, GOODACRE S, MASTA S E, et al.. Can long-range PCR be used to amplify genetically divergent mitochondrial genomes for comparative phylogenetics? A case study within spiders (Arthropoda: Araneae)[J/OL]. PLoS One, 2013, 8(5): e62404[2024-05-03]. . |
| 23 | JENSEN M R, SIGSGAARD E E, LIU S, et al.. Genome-scale target capture of mitochondrial and nuclear environmental DNA from water samples[J]. Mol. Ecol. Resour., 2021, 21(3): 690-702. |
| [1] | Ju CHEN, Chunjie DING, Yinling SUN, Han ZHAO, Hongyu ZHENG, Xin GAO, Shuhan JIN, Kun DONG. Antioxidant Activity and Composition Changes of Lycium chinense Miller Leaf Black Tea Fermented by Aspergillus cristatum [J]. Current Biotechnology, 2025, 15(3): 510-517. |
| [2] | Mengmeng KONG, Peng LU, Qiansi CHEN, Xueyi QIAO, Shanyi CHEN, Jingjing JIN, Xueao ZHENG, Peijian CAO, Jiemeng TAO. Construction of Cellulase Producing Engineering Strains and the Application in Aged Tobacco Leaves [J]. Current Biotechnology, 2024, 14(2): 263-270. |
| [3] | Xiao ZHANG, Caihua LI, Jing WANG, Lanlan ZHANG, Xiaoyu MOU, Xinyue WANG, Liumei GAN, Pengzhan ZHOU, Rui ZHANG. Impact of Gene Duplication on RNA Editing Rates of Mitochondrial atpA Gene in Cotton (Gossypium hirsutum L.) [J]. Current Biotechnology, 2022, 12(5): 737-745. |
| [4] | ZHAO Yu-fen1, LI Pei-yan2,3*, ZHENG Xiao-lin3, YIN Fei4. Absorption and Desorption of Total Flavonoids from Sweet Potato Leaves with Macroporous Resins [J]. Curr. Biotech., 2013, 3(6): 433-438. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||