Current Biotechnology ›› 2022, Vol. 12 ›› Issue (6): 880-887.DOI: 10.19586/j.2095-2341.2022.0084
• Articles • Previous Articles Next Articles
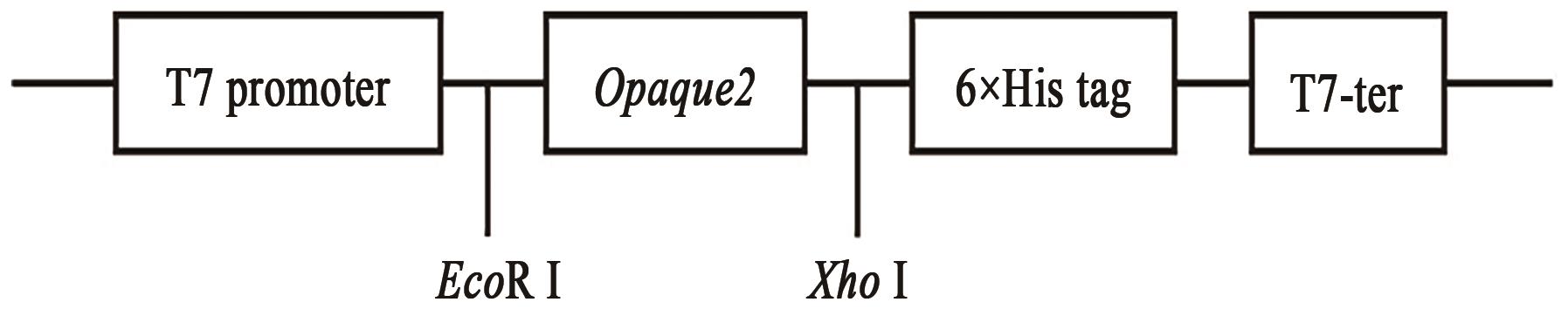
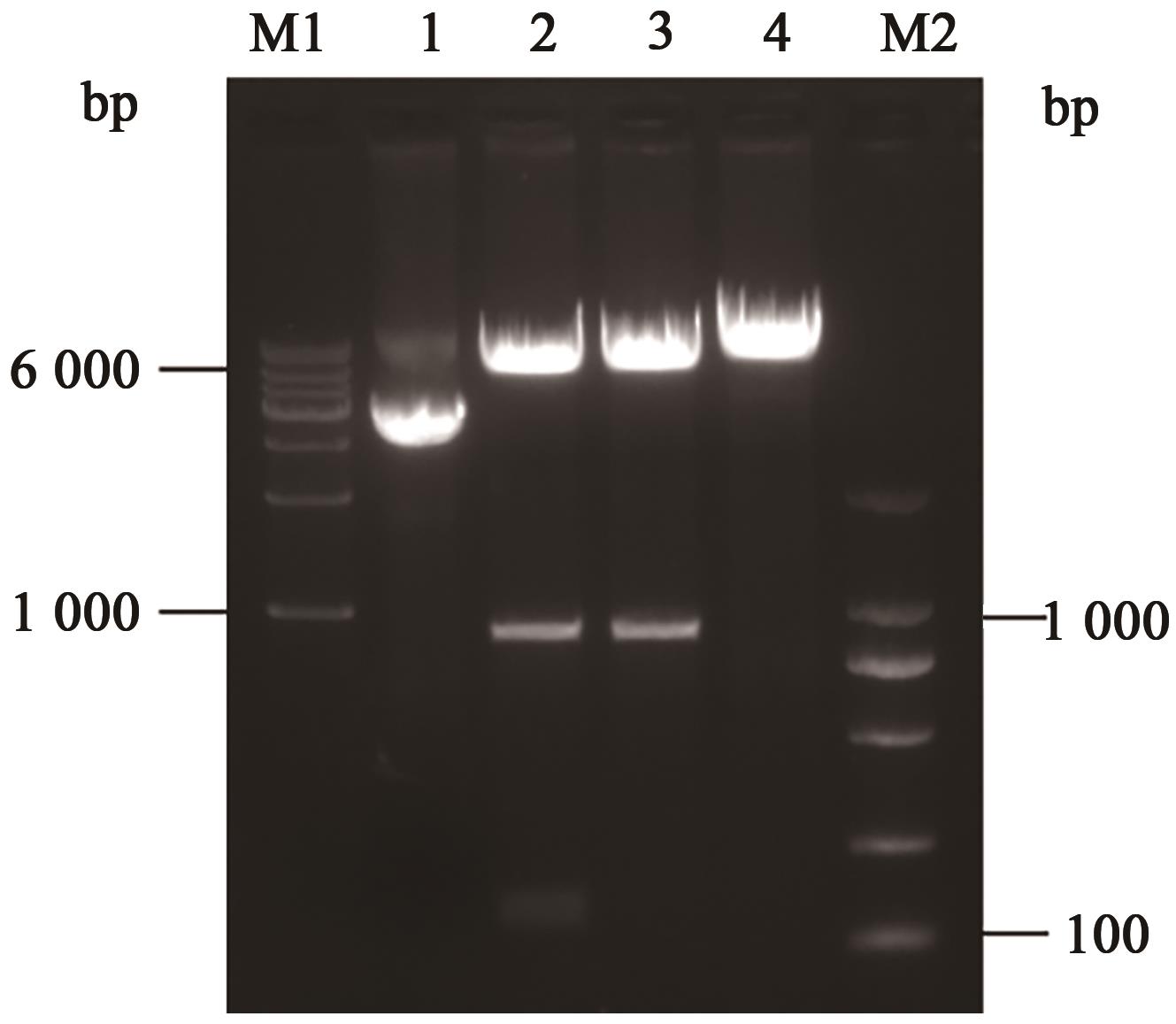
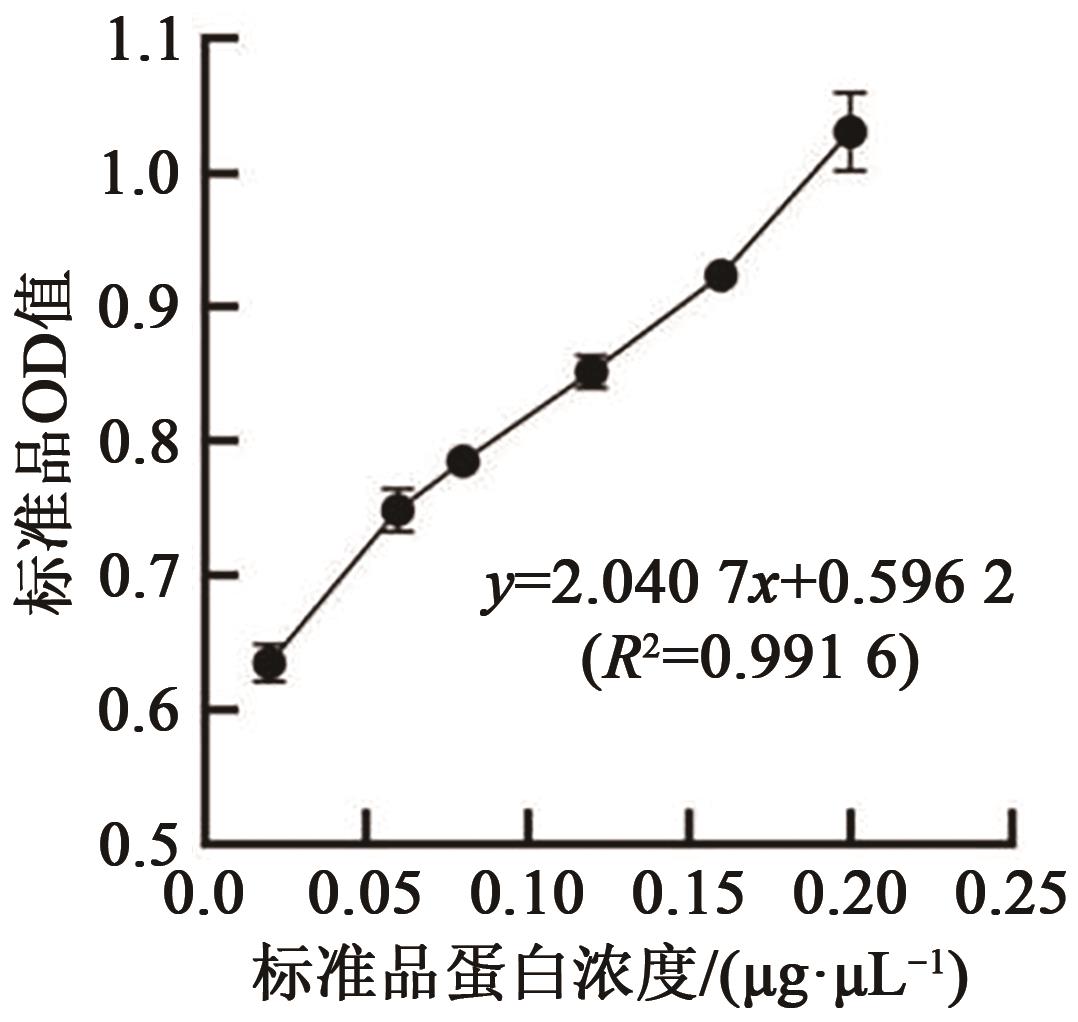
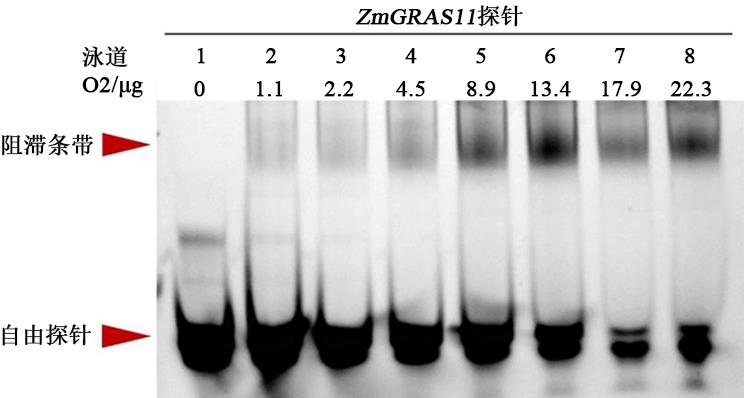
EMSA Experiments with Fluorescently Labeled Probes to Analyze the Binding Sites of Opaque2 Protein with the ZmGRAS11 Promoter
Jiameng ZHU1,2( ), Haiyang JIANG1, Rumei CHEN2, Xiaojin ZHOU2(
), Haiyang JIANG1, Rumei CHEN2, Xiaojin ZHOU2( )
)
- 1.School of Life Science,Anhui Agricultural University,Hefei 230000
2.Institute of Biotechnology,Chinese Academy of Agricultural Sciences,Beijing 100081
-
Received:2022-05-24Accepted:2022-06-30Online:2022-11-25Published:2022-11-30 -
Contact:Xiaojin ZHOU
通过荧光标记的凝胶阻滞技术分析Opaque2蛋白与ZmGRAS11启动子的结合位点
- 1.安徽农业大学生命科学学院,合肥 230000
2.中国农业科学院生物技术研究所,北京 100081
-
通讯作者:周晓今 -
作者简介:朱佳梦 E-mail: zhujiameng2021@163.com; -
基金资助:国家重点研发计划项目(2021YFF1000304)
CLC Number:
Cite this article
Jiameng ZHU, Haiyang JIANG, Rumei CHEN, Xiaojin ZHOU. EMSA Experiments with Fluorescently Labeled Probes to Analyze the Binding Sites of Opaque2 Protein with the ZmGRAS11 Promoter[J]. Current Biotechnology, 2022, 12(6): 880-887.
朱佳梦, 江海洋, 陈茹梅, 周晓今. 通过荧光标记的凝胶阻滞技术分析Opaque2蛋白与ZmGRAS11启动子的结合位点[J]. 生物技术进展, 2022, 12(6): 880-887.
share this article
| 试剂名称 | 配方 | pH |
|---|---|---|
| 1 mol·L-1 IPTG | 2.38301 g IPTG,10 mL 超纯水 | — |
| 裂解缓冲液 | 0.02 mol·L-1 咪唑,0.02 mol·L-1 Tris,0.5 mol·L-1 氯化钠,10% 甘油,0.5% TritonX-100 | 8.0 |
| 洗杂缓冲液A | 0.02 mol·L-1咪唑,0.02 mol·L-1 Tris,0.5 mol·L-1 氯化钠,10% 甘油,0.5% TritonX-100 | 8.0 |
| 洗杂缓冲液B | 0.04 mol·L-1咪唑,0.02 mol·L-1 Tris,0.5 mol·L-1 氯化钠,10%甘油,0.5% TritonX-100 | 8.0 |
| 洗杂缓冲液C | 0.08 mol·L-1 咪唑,0.02 mol·L-1 Tris,0.5 mol·L-1 氯化钠,10% 甘油,0.5% TritonX-100 | 8.0 |
| 洗脱缓冲液 | 0.25 mol·L-1咪唑,0.02 mol·L-1 Tris,0.5 mol·L-1 氯化钠,10% 甘油 | 8.0 |
| 1×置换缓冲液 | 0.05 mol·L-1 Tris,0.25 mol·L-1 氯化钠,0.005 mol·L-1 DTT | 7.5 |
| 5×TBE缓冲液 | 0.45 mol·L-1 Tris,0.45 mol·L-1 硼酸,0.01 mol·L-1 EDTA | 8.3 |
Table 1 Formulation of reagent used in this study
| 试剂名称 | 配方 | pH |
|---|---|---|
| 1 mol·L-1 IPTG | 2.38301 g IPTG,10 mL 超纯水 | — |
| 裂解缓冲液 | 0.02 mol·L-1 咪唑,0.02 mol·L-1 Tris,0.5 mol·L-1 氯化钠,10% 甘油,0.5% TritonX-100 | 8.0 |
| 洗杂缓冲液A | 0.02 mol·L-1咪唑,0.02 mol·L-1 Tris,0.5 mol·L-1 氯化钠,10% 甘油,0.5% TritonX-100 | 8.0 |
| 洗杂缓冲液B | 0.04 mol·L-1咪唑,0.02 mol·L-1 Tris,0.5 mol·L-1 氯化钠,10%甘油,0.5% TritonX-100 | 8.0 |
| 洗杂缓冲液C | 0.08 mol·L-1 咪唑,0.02 mol·L-1 Tris,0.5 mol·L-1 氯化钠,10% 甘油,0.5% TritonX-100 | 8.0 |
| 洗脱缓冲液 | 0.25 mol·L-1咪唑,0.02 mol·L-1 Tris,0.5 mol·L-1 氯化钠,10% 甘油 | 8.0 |
| 1×置换缓冲液 | 0.05 mol·L-1 Tris,0.25 mol·L-1 氯化钠,0.005 mol·L-1 DTT | 7.5 |
| 5×TBE缓冲液 | 0.45 mol·L-1 Tris,0.45 mol·L-1 硼酸,0.01 mol·L-1 EDTA | 8.3 |
| 试管号 | 菌液加入量/mL | IPTG加入量/μL | IPTG诱导浓度/(mmol·L-1) |
|---|---|---|---|
| a | 6 | 0 | 0 |
| b | 6 | 1.2 | 0.2 |
| c | 6 | 2.4 | 0.4 |
| d | 6 | 3.6 | 0.6 |
| e | 6 | 4.8 | 0.8 |
| f | 6 | 6.0 | 1.0 |
Table 2 Optimization of IPTG concentration for recombinant protein expression
| 试管号 | 菌液加入量/mL | IPTG加入量/μL | IPTG诱导浓度/(mmol·L-1) |
|---|---|---|---|
| a | 6 | 0 | 0 |
| b | 6 | 1.2 | 0.2 |
| c | 6 | 2.4 | 0.4 |
| d | 6 | 3.6 | 0.6 |
| e | 6 | 4.8 | 0.8 |
| f | 6 | 6.0 | 1.0 |
| 探针名称 | 序列(5'→3') | 标记 |
|---|---|---|
| 5'-GGCACTAGTCATGCTAGCTTGTGACTCATCCAACAAACAATAGTTATCC-3' | 5'-FAM | |
| FAM-R | 5'-GGATAACTATTGTTTGTTGGATGAGTCACAAGCTAGCATGACTAGTGCC-3' | 5'-FAM |
| WT-F | 5'-GGCACTAGTCATGCTAGCTTGTGACTCATCCAACAAACAATAGTTATCC-3' | — |
| WT-F | 5'-GGATAACTATTGTTTGTTGGATGAGTCACAAGCTAGCATGACTAGTGCC-3' | — |
| M1-F | 5'-GGCACTAGTCATGCTAGCTTGTCCAACAAACAATAGTTATCC-3' | — |
| M1-R | 5'-GGATAACTATTGTTTGTTGGACAAGCTAGCATGACTAGTGCC-3' | — |
| M2-F | 5'-GGCACTAGTCATGCTAGCTTGTTTTTTTTCCAACAAACAATAGTTATCC-3' | — |
| M2-R | 5'-GGATAACTATTGTTTGTTGGAAAAAAAACAAGCTAGCATGACTAGTGCC-3' | — |
| M3-F | 5'-GGCACTAGTCATGCTAGCTTGGGGGGGGTCCAACAAACAATAGTTATCC-3' | — |
| M3-R | 5'-GGATAACTATTGTTTGTTGGACCCCCCCCAAGCTAGCATGACTAGTGCC-3' | — |
| M4-F | 5'-GGCACTAGTCATGCTAGCTTGCAGCTCATCCAACAAACAATAGTTATCC-3' | — |
| M4-R | 5'-GGATAACTATTGTTTGTTGGATGAGCTGCAAGCTAGCATGACTAGTGCC-3' | — |
| M5-F | 5'-GGCACTAGTCATGCTAGCTTGTGGTCCATCCAACAAACAATAGTTATCC-3' | — |
| M5-R | 5'-GGATAACTATTGTTTGTTGGATGGACCACAAGCTAGCATGACTAGTGCC-3' | — |
| M6-F | 5'-GGCACTAGTCATGCTAGCTTGTGACCTGTCCAACAAACAATAGTTATCC-3' | — |
| M6-R | 5'-GGATAACTATTGTTTGTTGGACAGGTCACAAGCTAGCATGACTAGTGC-3' | — |
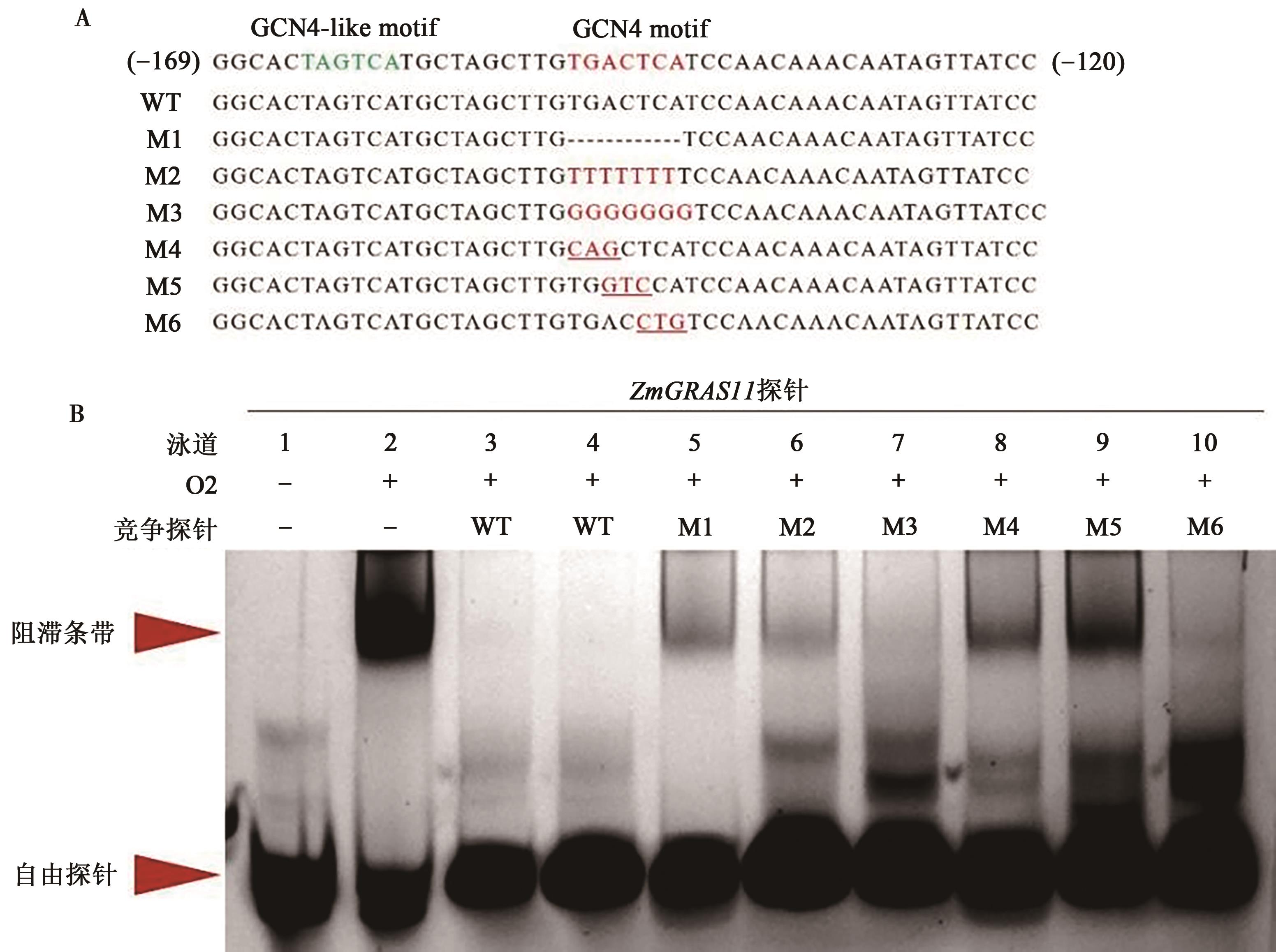
Table 3 Probes in the ZmGRAS11 promoter
| 探针名称 | 序列(5'→3') | 标记 |
|---|---|---|
| 5'-GGCACTAGTCATGCTAGCTTGTGACTCATCCAACAAACAATAGTTATCC-3' | 5'-FAM | |
| FAM-R | 5'-GGATAACTATTGTTTGTTGGATGAGTCACAAGCTAGCATGACTAGTGCC-3' | 5'-FAM |
| WT-F | 5'-GGCACTAGTCATGCTAGCTTGTGACTCATCCAACAAACAATAGTTATCC-3' | — |
| WT-F | 5'-GGATAACTATTGTTTGTTGGATGAGTCACAAGCTAGCATGACTAGTGCC-3' | — |
| M1-F | 5'-GGCACTAGTCATGCTAGCTTGTCCAACAAACAATAGTTATCC-3' | — |
| M1-R | 5'-GGATAACTATTGTTTGTTGGACAAGCTAGCATGACTAGTGCC-3' | — |
| M2-F | 5'-GGCACTAGTCATGCTAGCTTGTTTTTTTTCCAACAAACAATAGTTATCC-3' | — |
| M2-R | 5'-GGATAACTATTGTTTGTTGGAAAAAAAACAAGCTAGCATGACTAGTGCC-3' | — |
| M3-F | 5'-GGCACTAGTCATGCTAGCTTGGGGGGGGTCCAACAAACAATAGTTATCC-3' | — |
| M3-R | 5'-GGATAACTATTGTTTGTTGGACCCCCCCCAAGCTAGCATGACTAGTGCC-3' | — |
| M4-F | 5'-GGCACTAGTCATGCTAGCTTGCAGCTCATCCAACAAACAATAGTTATCC-3' | — |
| M4-R | 5'-GGATAACTATTGTTTGTTGGATGAGCTGCAAGCTAGCATGACTAGTGCC-3' | — |
| M5-F | 5'-GGCACTAGTCATGCTAGCTTGTGGTCCATCCAACAAACAATAGTTATCC-3' | — |
| M5-R | 5'-GGATAACTATTGTTTGTTGGATGGACCACAAGCTAGCATGACTAGTGCC-3' | — |
| M6-F | 5'-GGCACTAGTCATGCTAGCTTGTGACCTGTCCAACAAACAATAGTTATCC-3' | — |
| M6-R | 5'-GGATAACTATTGTTTGTTGGACAGGTCACAAGCTAGCATGACTAGTGC-3' | — |
| 各个组分加入量/(μL) | 泳道 | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| 超纯水 | 14.0 | 13.5 | 13.0 | 12.0 | 10.0 | 8.0 | 6.0 | 4.0 |
| 5×结合缓冲液 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| O2蛋白 | 0 | 0.5 | 1.0 | 2.0 | 4.0 | 6.0 | 8.0 | 10.0 |
Table 4 Different treatments of protein to probe ratio
| 各个组分加入量/(μL) | 泳道 | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| 超纯水 | 14.0 | 13.5 | 13.0 | 12.0 | 10.0 | 8.0 | 6.0 | 4.0 |
| 5×结合缓冲液 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| O2蛋白 | 0 | 0.5 | 1.0 | 2.0 | 4.0 | 6.0 | 8.0 | 10.0 |
| 各个组分加入量/(μL) | 泳道 | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| 超纯水 | 14 | 10 | 5 | 0 | 0 | 0 | 0 | 0 |
| 5×结合缓冲液 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| O2蛋白 | 0 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| 竞争探针 | 0 | 0 | 5 | 10 | 10 | 10 | 10 | 10 |
Table 5 Reaction system of competitive probes with different mutations
| 各个组分加入量/(μL) | 泳道 | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| 超纯水 | 14 | 10 | 5 | 0 | 0 | 0 | 0 | 0 |
| 5×结合缓冲液 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| O2蛋白 | 0 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| 竞争探针 | 0 | 0 | 5 | 10 | 10 | 10 | 10 | 10 |
| 1 | SITARAMAYYA A, WRIGHT L S, SIIEGEL F L. Enzymatic methylation of calmodulin in rat brain cytosol[J]. Biol. Chem., 1980, 255(18): 8894-8900. |
| 2 | HOWE C L, MOOSEKER M S, GRAVES T A. Brush-border calmodulin. A major component of the isolated microvillus core[J]. J.Cell Biol., 1980, 85: 916-923. |
| 3 | CHELM B K, GEIDUSCHEK E P. Gel electrophoretic separation of transcription complexes: an assay for RNA polymerase selectivity and a method for promoter mapping[J]. Nucl. Acids Res., 1979, 7(7): 1851-1867. |
| 4 | VARSHAVSKY A J, BAKAYEV V V, GEORGIEV G P. Heterogeneity of chromatin subunits in vitro and location of histone H1[J]. Nucl. Acids Res., 1976, 3(2): 477-492. |
| 5 | RAMANATHAN M, PORTER D F, KHAVARI P A. Methods to study RNA-protein interactions[J]. Nat. Methods., 2019, 16(3): 225-234. |
| 6 | REAM J A, LEWIS L K, LEWIS K A. Rapid agarose gel electrophoretic mobility shift assay for quantitating protein: RNA interactions[J]. Anal. Biochem., 2016, 511: 36-41. |
| 7 | SEO M, LEI L, EGLI M. Label-free electrophoretic mobility shift assay (EMSA) for measuring dissociation constants of protein-RNA complexes[J/OL]. Curr. Protoc. Nucl. Acid Chem., 2019, 76(1): e70[2022-06-01]. . |
| 8 | MAXAM A, GILBERT W S. A new method for sequencing DNA[J]. Proc. Natl. Acad. Sci. USA, 1977, 74: 560-565. |
| 9 | HELLMAN L M, FRIED M G. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions[J]. Nat. Protoc., 2007, 2(8): 1849-1861. |
| 10 | TIAN J, WANG C, XIA J, et al.. Teosinte ligule allele narrows plant architecture and enhances high-density maize yields[J]. Science, 2019, 365(6454): 658-664. |
| 11 | KONG D, PAN X, JING Y, et al.. ZmSPL10/14/26 are required for epidermal hair cell fate specification on maize leaf[J]. New Phytol., 2021, 230(4): 1533-1549. |
| 12 | XIE Y, LIU Y, WANG H, et al.. Phytochrome-interacting factors directly suppress MIR156 expression to enhance shade-avoidance syndrome in Arabidopsis [J]. Nat. Commun., 2017, 8(1): 1-11. |
| 13 | LI C, SONG R. The regulation of zein biosynthesis in maize endosperm[J]. Theor. Appl. Genet., 2020, 133(5): 1443-1453. |
| 14 | LI C, YUE Y, CHEN H, et al.. The ZmbZIP22 transcription factor regulates 27-kD γ-Zein gene transcription during maize endosperm development[J]. Plant Cell, 2018, 30(10): 2402-2424. |
| 15 | QIAO Z, QI W, WANG Q, et al.. ZmMADS47 regulates Zein gene transcription through interaction with Opaque2[J/OL]. PLoS Genet., 2016, 12(4): e1005991[2022-06-01]. . |
| 16 | VICENTE-CARBAJOSA J, MOOSE S P, PARSONS R L, et al.. A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2[J]. Proc. Natl. Acad. Sci. USA, 1997, 94(14): 7685-7690. |
| 17 | DENG Y, WANG J, ZHANG Z, et al.. Transactivation of Sus1 and Sus2 by Opaque2 is an essential supplement to sucrose synthase-mediated endosperm filling in maize[J]. Plant Biotechnol. J., 2020, 18(9): 1897-1907. |
| 18 | ZHOU Z, SONG L, ZHANG X, et al.. Introgression of opaque2 into waxy maize causes extensive biochemical and proteomic changes in endosperm[J/OL]. PLoS ONE, 2016, 11(7): e0158971[2022-06-01]. . |
| 19 | LI Y, MA S, ZHAO Q, et al.. ZmGRAS11, transactivated by Opaque2, positively regulates kernel size in maize[J]. J. Integr. Plant Biol., 2021, 63(12): 2031-2037. |
| 20 | GAO Y, AN K, GUO W, et al.. The endosperm-specific transcription factor TaNAC019 regulates glutenin and starch accumulation and its elite allele improves wheat grain quality[J]. Plant Cell, 2021, 33(3): 603-622. |
| 21 | LI C, QIAO Z, QI W, et al.. Genome-wide characterization of cis-acting DNA targets reveals the transcriptional regulatory framework of opaque2 in maize[J]. Plant Cell, 2015, 27(3): 532-545. |
| 22 | LIU Y, MA M, LI G, et al.. Transcription factors FHY3 and FAR1 regulate light-induced CIRCADIAN CLOCK ASSOCIATED1 gene expression in Arabidopsis [J]. Plant Cell, 2020, 32(5): 1464-1478. |
| 23 | ZONG W, TANG N, YANG J, et al.. Feedback regulation of ABA signaling and biosynthesis by a bZIP transcription factor targets drought-resistance-related genes[J]. Plant Physiol., 2016, 171(4): 2810-2825. |
| 24 | 刘蕾,周欣悦,朱桢,等. 副溶血弧菌外膜铁蛋白受体pvuA基因的原核表达及产物的诱导条件优化[J].生物技术进展, 2022,12(3):396-404. |
| 25 | 陈威风,崔丽伟,常惟丹,等.金黄色葡萄球菌肠毒素A、B、C、D、E重组载体的构建及表达[J].生物技术进展, 2022,12(2):313-317. |
| 26 | GALHANO R, ILLANA A, RYDER L S, et al.. Tpc1 is an important Zn(II)2Cys6 transcriptional regulator required for polarized growth and virulence in the rice blast fungus[J/OL]. PLoS Pathog., 2017, 13(7): e1006516[2022-06-01]. . |
| 27 | ZHANG Z, ZHENG X, YANG J, et al.. Maize endosperm-specific transcription factors O2 and PBF network the regulation of protein and starch synthesis[J]. Proc. Natl. Acad. Sci. USA, 2016, 113(39): 10842-10847. |
| 28 | SHPJI T, HASHIMOTO T. Tobacco MYC2 regulates jasmonate-inducible nicotine biosynthesis genes directly and by way of the NIC2-locus ERF genes[J]. Plant Cell Physiol., 2011, 52(6): 1117-1130. |
| 29 | MENG Y, WANG Z, WANG Y, et al.. The MYB activator WHITE PETAL1 associates with MtTT8 and MtWD40-1 to regulate carotenoid-derived flower pigmentation in Medicago truncatula [J]. Plant Cell, 2019, 31(11): 2751-2767. |
| 30 | JI C, XU L, LI Y, et al.. The O2-ZmGRAS11 transcriptional regulatory network orchestrates the coordination of endosperm cell expansion and grain filling in maize[J]. Mol. Plant., 2022, 15(3): 468-487. |
| [1] | Zhenhua XU, Shiwei GAO, Dawei GAO, Yanming YU, Haiying LIU, Hongtao WU, Shuli ZHANG, Zhongyi SUN, Xin WANG, Ping YAN. Biological Breeding Science and Technology Innovation for Seed Industry Development and Prospects [J]. Current Biotechnology, 2025, 15(4): 557-564. |
| [2] | Wanwan LYU, Lin ZHENG, Hongying PANG, Hongbo GAO, Hongzhi WANG. Research Advances on the Mechanism of Root Suckering in Poplar [J]. Current Biotechnology, 2025, 15(3): 372-379. |
| [3] | Zhuoying LIU, Xiaojin ZHOU, Yanli HUANG, Sen PANG. Joint Transcriptome Analysis of Maize Under Salt Stress and MeJA Treatment [J]. Current Biotechnology, 2025, 15(2): 263-275. |
| [4] | Lanlan ZHANG, Caihua LI, Yuzhu FANG, Yan SONG, Wanlin KANG, Zhiyu LI, Xiao ZHANG, Rui ZHANG. Progress on the Application of Mitochondrial SSR Molecular Markers in Plants [J]. Current Biotechnology, 2023, 13(6): 821-826. |
| [5] | Min LI, Lei WANG, Junjie ZOU. Opportunities and Challenges for the Industrial Application of Transgenic Insect-resistant and Herbicide-tolerant Maize in China [J]. Current Biotechnology, 2023, 13(2): 157-165. |
| [6] | Hui SUN, Chunyi ZHANG, Ling JIANG. Progress of Plant Molecular Farming in Pharmaceutical Use [J]. Current Biotechnology, 2023, 13(1): 65-71. |
| [7] | Yang YANG, Fenglin WANG, De LIU, Yuanyuan LUO, Jianhua ZHU. Research Progress of CRISPR⁃Cas9 Technology on the Production of Plant Secondary Metabolites [J]. Current Biotechnology, 2022, 12(6): 806-816. |
| [8] | Zhaohui QIAO, Liang CHEN. Dynamic Regulation and Physiological Role of Promoter-proximal Pause/Release of RNA Polymerase Ⅱ: a Review [J]. Current Biotechnology, 2022, 12(5): 705-710. |
| [9] | Qin ZHOU, Yurong XIE. Functional Analysis of ELONGATED HYPOCOTYL5 Regulating Branching in Arabidopsis thaliana [J]. Current Biotechnology, 2022, 12(3): 379-386. |
| [10] | Yunyan FEI, Jun YANG, Dedao JING, Tianzi LIN, Chuang LI, Huafei QIAN, Shengyuan ZENG, Huaxin HAN, Hongbing GONG. Research and Application Progress of CRISPR/Cas Technology in Herbicide⁃resistant Crops Breeding [J]. Current Biotechnology, 2022, 12(2): 189-197. |
| [11] | Jiu HUANG, Shuangfeng SHI, Erte ZHANG, Mei LI, Yue YU, Yuhui LIU. Cloning and Functional Studies of Nitrate Transporter Gene GeNRT2.1 in Glossostigma elatinoides [J]. Current Biotechnology, 2022, 12(2): 256-264. |
| [12] | Xin WANG, Tianzhu ZHANG. Research Progress on the Regulation of Anthocyanin Synthesis in Horticultural Crops [J]. Current Biotechnology, 2022, 12(1): 10-16. |
| [13] | Huilu XU, Min XU, Wei ZHANG. The Effect of Deleting the slr1556 Gene Encoding Lactate Dehydrogenase on Ethanol Biosynthesize in Synechocystis sp. PCC 6803 [J]. Current Biotechnology, 2022, 12(1): 105-111. |
| [14] | Mengjie LI, Xiaoyun HE, Tao TONG, Jingang LIANG, Kunlun HUANG. Overview and Progress of Safety Regulatory System of Genetically Modified Crops in Argentina [J]. Current Biotechnology, 2021, 11(6): 676-687. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||