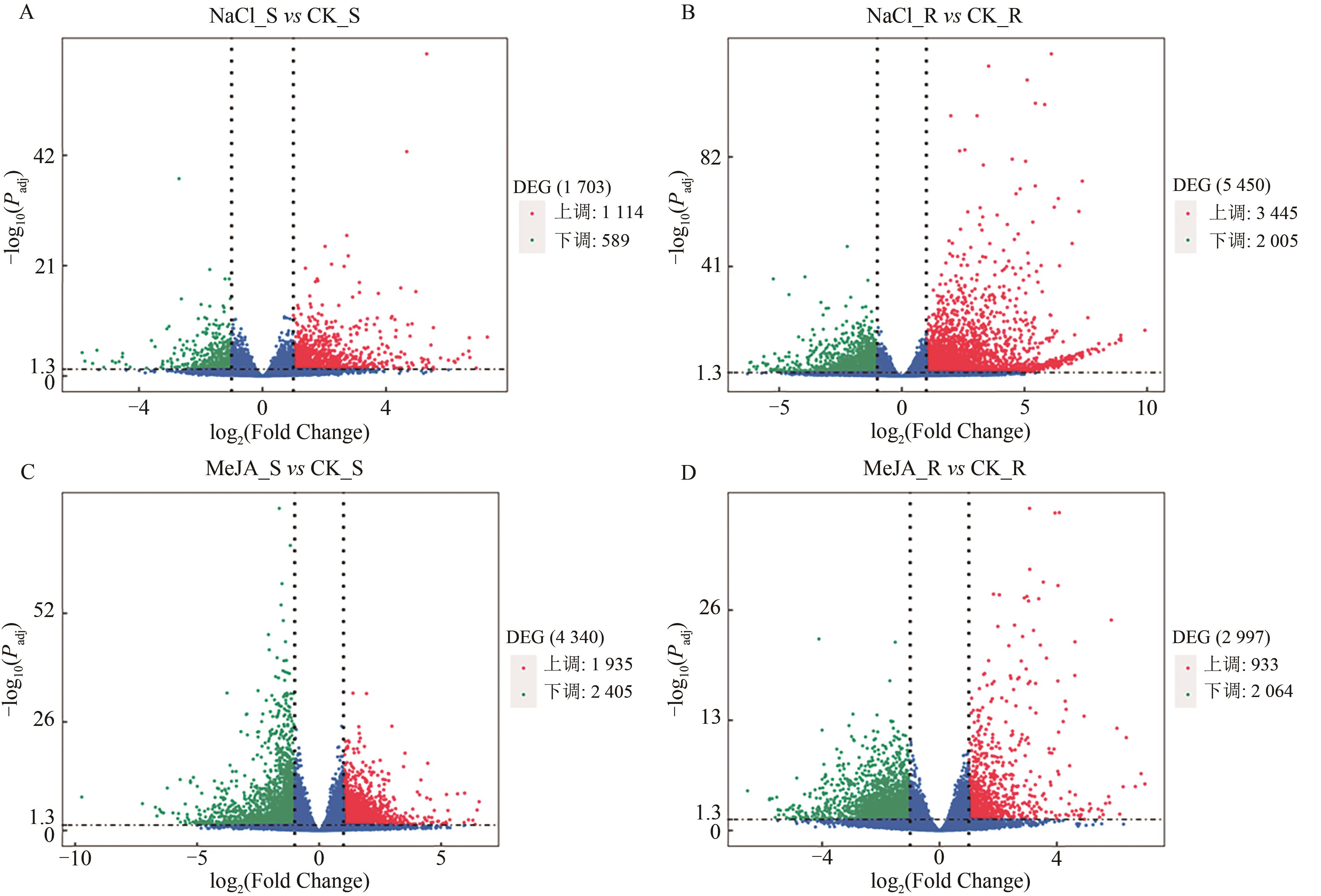
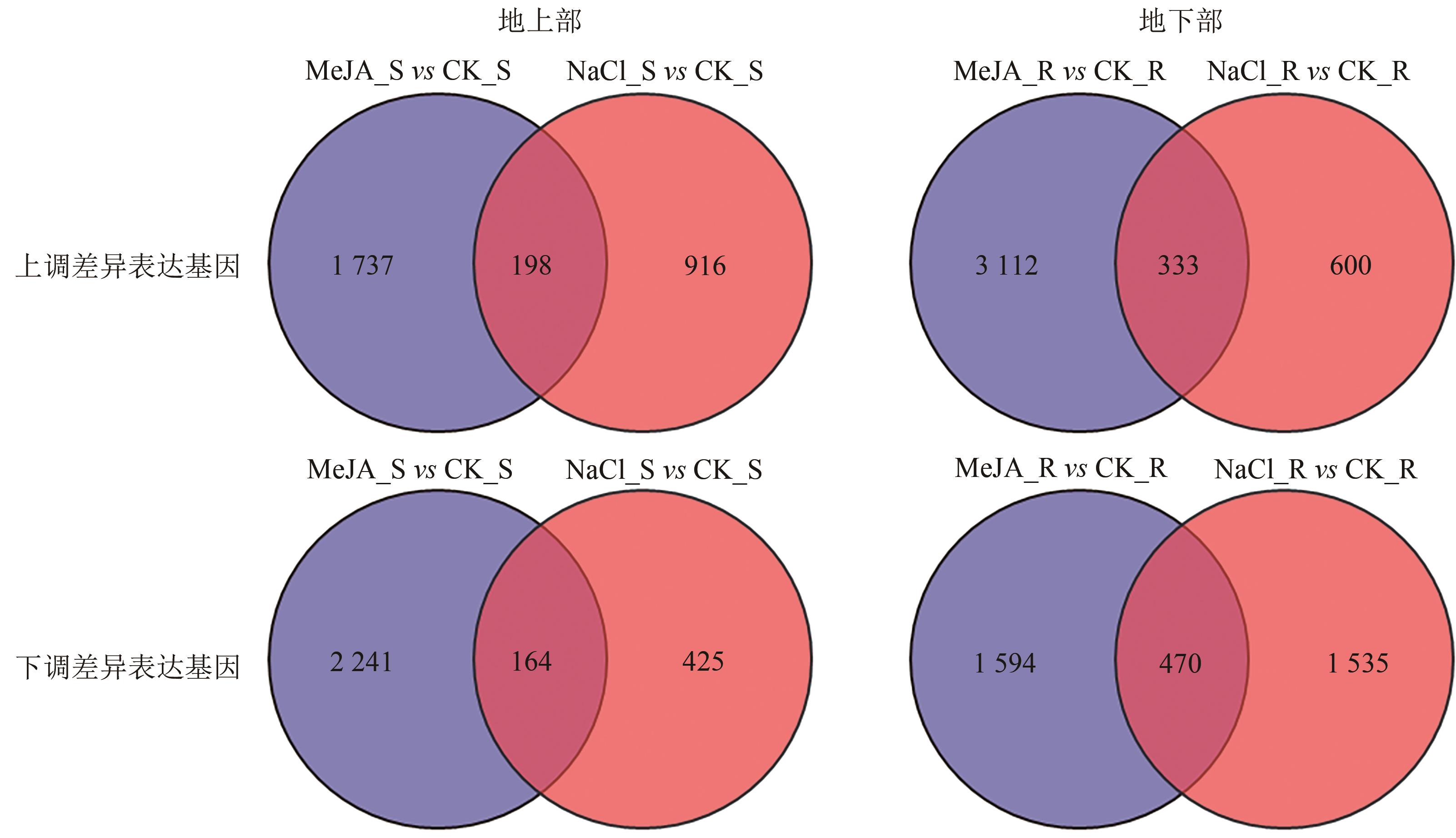
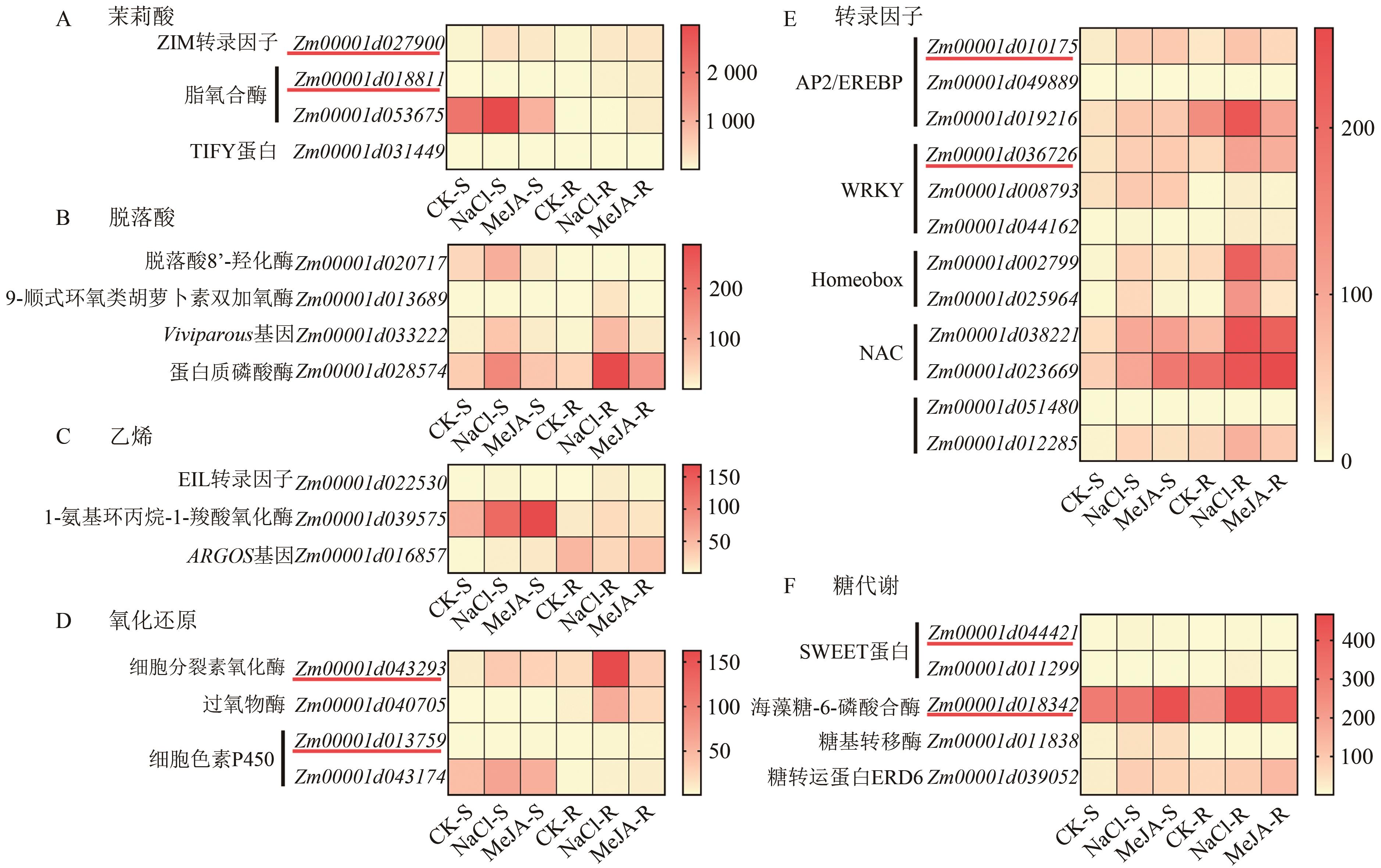
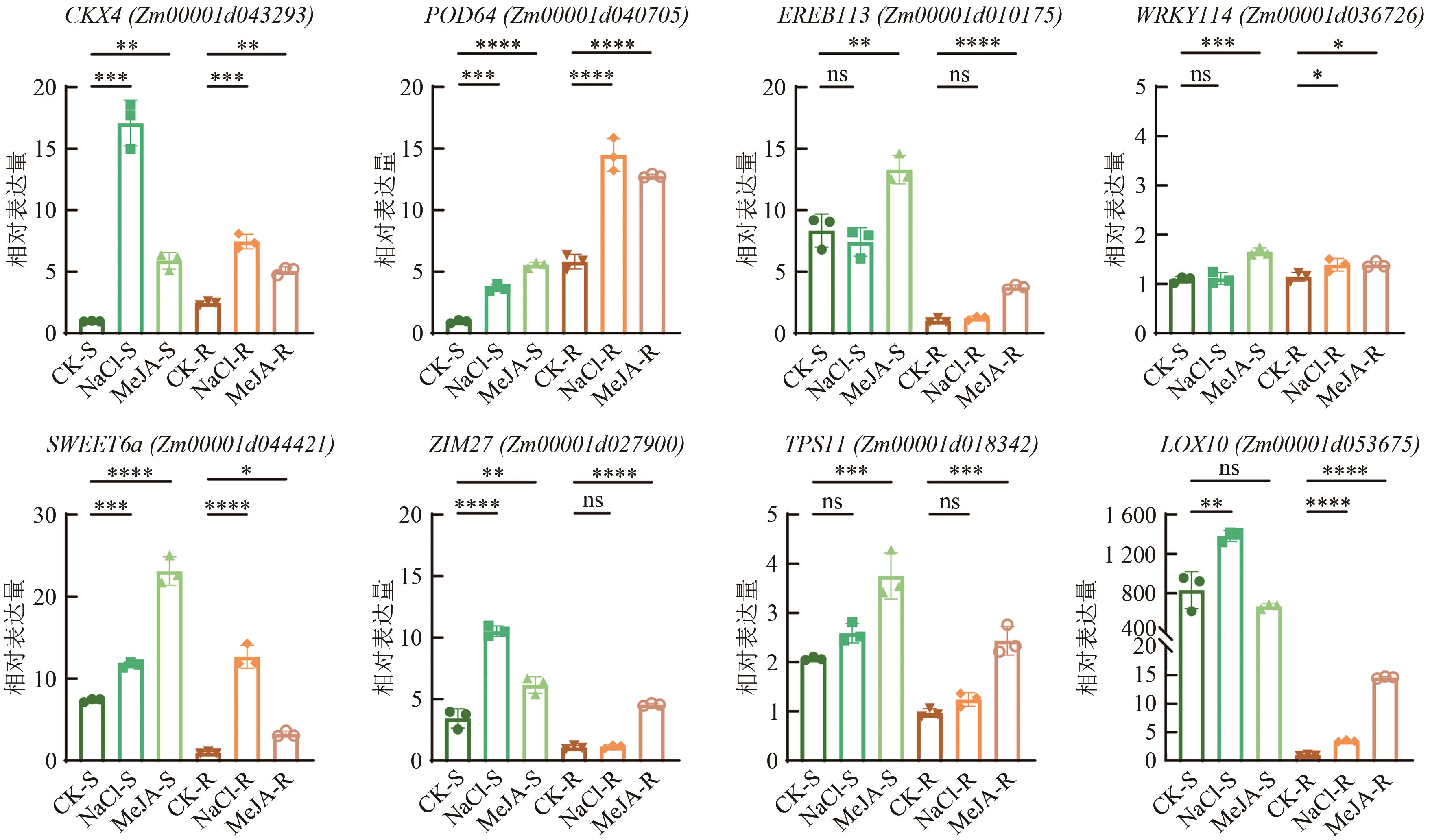
Current Biotechnology ›› 2025, Vol. 15 ›› Issue (2): 263-275.DOI: 10.19586/j.2095-2341.2024.0183
• Articles • Previous Articles Next Articles
Joint Transcriptome Analysis of Maize Under Salt Stress and MeJA Treatment
Zhuoying LIU1( ), Xiaojin ZHOU2(
), Xiaojin ZHOU2( ), Yanli HUANG3, Sen PANG1(
), Yanli HUANG3, Sen PANG1( )
)
- 1.College of Science,China Agricultural University,Beijing 100193,China
2.Biotechnology Research Institute,Chinese Academy of Agricultural Sciences,Beijing 100081,China
3.School of Innovation and Entrepreneurship,Heze University,Shandong Heze 274000,China
-
Received:2024-11-24Accepted:2025-01-21Online:2025-03-25Published:2025-04-29 -
Contact:Xiaojin ZHOU,Sen PANG
玉米盐胁迫和MeJA处理下的转录组联合分析
- 1.中国农业大学理学院,北京 100193
2.中国农业科学院生物技术研究所,北京 100081
3.菏泽学院创新创业学院,山东 菏泽 274000
-
通讯作者:周晓今,逄森 -
作者简介:刘卓颖E-mail:zz9629@126.com -
基金资助:国家自然科学基金面上项目(32472217)
CLC Number:
Cite this article
Zhuoying LIU, Xiaojin ZHOU, Yanli HUANG, Sen PANG. Joint Transcriptome Analysis of Maize Under Salt Stress and MeJA Treatment[J]. Current Biotechnology, 2025, 15(2): 263-275.
刘卓颖, 周晓今, 黄燕丽, 逄森. 玉米盐胁迫和MeJA处理下的转录组联合分析[J]. 生物技术进展, 2025, 15(2): 263-275.
share this article
| 基因名称(ID) | 上游引物序列(5'→3') | 下游引物序列(5'→3') |
|---|---|---|
| ZmActin (Zm00001d013410) | 5'-ATGTTTCCTGGGATTGCCGAT-3' | 5'-CCAGTTTCGTCATACTCTCCCTTG-3' |
| SWEET6a (Zm00001d044421) | 5'-CGAGACGTGCTTTACCCACA-3' | 5'-GAATGGCAAACACAGCCACA-3' |
| CKX4 (Zm00001d043293) | 5'-CGGCCGACTCGTAACGTAAT-3' | 5'-CACGCGATTAACAACCCCAC-3' |
| POD64 (Zm00001d040705) | 5'-GTGCCCAACTCCTACTCTGG-3' | 5'-TGACACACGAACTACGCACT-3' |
| EREB113 (Zm00001d010175) | 5'-GCCAAGGCAGCAGCAGTCA-3' | 5'-GGCACAGAAGCCACGGTAA-3' |
| WRKY114 (Zm00001d036726) | 5'-CTAGCTCTAGCACGTACGCC-3' | 5'-GAGCGGTACAAACTCGGTCA-3' |
| ZIM27 (Zm00001d027900) | 5'-AGGAAGGTGTCGCTAAAGAG-3' | 5'-CTGCCGTCGATGAGATTG-3' |
| TPS11 (Zm00001d018342) | 5'-CGGCGTGGTGTAGGATTGAT-3' | 5'-ACCATGCGGGCATATACAGG-3' |
| LOX10 (Zm00001d053675) | 5'-TTCCAAACAGCATCTCCATT-3' | 5'-GCCTTATTACAACAGTCCTCACG-3' |
Table 1 Primer sequences used for qPCR analysis
| 基因名称(ID) | 上游引物序列(5'→3') | 下游引物序列(5'→3') |
|---|---|---|
| ZmActin (Zm00001d013410) | 5'-ATGTTTCCTGGGATTGCCGAT-3' | 5'-CCAGTTTCGTCATACTCTCCCTTG-3' |
| SWEET6a (Zm00001d044421) | 5'-CGAGACGTGCTTTACCCACA-3' | 5'-GAATGGCAAACACAGCCACA-3' |
| CKX4 (Zm00001d043293) | 5'-CGGCCGACTCGTAACGTAAT-3' | 5'-CACGCGATTAACAACCCCAC-3' |
| POD64 (Zm00001d040705) | 5'-GTGCCCAACTCCTACTCTGG-3' | 5'-TGACACACGAACTACGCACT-3' |
| EREB113 (Zm00001d010175) | 5'-GCCAAGGCAGCAGCAGTCA-3' | 5'-GGCACAGAAGCCACGGTAA-3' |
| WRKY114 (Zm00001d036726) | 5'-CTAGCTCTAGCACGTACGCC-3' | 5'-GAGCGGTACAAACTCGGTCA-3' |
| ZIM27 (Zm00001d027900) | 5'-AGGAAGGTGTCGCTAAAGAG-3' | 5'-CTGCCGTCGATGAGATTG-3' |
| TPS11 (Zm00001d018342) | 5'-CGGCGTGGTGTAGGATTGAT-3' | 5'-ACCATGCGGGCATATACAGG-3' |
| LOX10 (Zm00001d053675) | 5'-TTCCAAACAGCATCTCCATT-3' | 5'-GCCTTATTACAACAGTCCTCACG-3' |
| 样本 | 原始Reads | 过滤后Reads | Q20 | Q30 | GC含量 | Uniquely mapped |
|---|---|---|---|---|---|---|
| CK-S-1 | 43678100 | 43047426 | 98.30 % | 95.06 % | 55.69 % | 95.74 % |
| CK-S-2 | 50852996 | 50083122 | 98.18 % | 94.75 % | 55.37 % | 95.44 % |
| CK-S-3 | 43444624 | 42661464 | 98.27 % | 94.99 % | 55.15 % | 95.64 % |
| CK-S-4 | 51020748 | 50012420 | 98.19 % | 94.90 % | 56.15 % | 95.19 % |
| CK-R-1 | 47340142 | 46607418 | 98.23 % | 94.92 % | 55.15 % | 93.16 % |
| CK-R-2 | 42318602 | 41636470 | 98.28 % | 95.09 % | 55.11 % | 95.27 % |
| CK-R-3 | 42916752 | 42228274 | 98.32 % | 95.14 % | 54.54 % | 93.34 % |
| CK-R-4 | 47196988 | 46286054 | 98.24 % | 94.94 % | 55.29 % | 93.58 % |
| NaCl-S-1 | 44346810 | 43693600 | 98.41 % | 95.37 % | 55.97 % | 95.90 % |
| NaCl-S-2 | 44482508 | 43736948 | 98.13 % | 94.64 % | 56.02 % | 95.60 % |
| NaCl-S-3 | 45904696 | 45070904 | 98.23 % | 94.89 % | 55.94 % | 96.16 % |
| NaCl-S-4 | 55913340 | 54947042 | 98.20 % | 94.86 % | 55.90 % | 95.91 % |
| NaCl-R-1 | 47132296 | 46300150 | 98.25 % | 95.00 % | 55.83 % | 93.94 % |
| NaCl-R-2 | 42765116 | 42001334 | 98.09 % | 94.64 % | 55.81 % | 93.33 % |
| NaCl-R-3 | 44430198 | 43679556 | 98.17 % | 94.79 % | 56.33 % | 93.88 % |
| NaCl-R-4 | 44579594 | 43831320 | 98.28 % | 95.08 % | 56.02 % | 94.16 % |
| MeJA-S-1 | 53707412 | 52824432 | 98.23 % | 94.92 % | 54.87 % | 94.11 % |
| MeJA-S-2 | 43478130 | 42516802 | 98.24 % | 94.97 % | 54.42 % | 94.95 % |
| MeJA-S-3 | 40559722 | 39809098 | 97.95 % | 94.15 % | 54.50 % | 95.22 % |
| MeJA-S-4 | 46413454 | 45685218 | 98.37 % | 95.28 % | 54.46 % | 95.43 % |
| MeJA-R-1 | 42939854 | 41593670 | 98.30 % | 95.02 % | 54.49 % | 88.28 % |
| MeJA-R-2 | 43131000 | 42474978 | 97.98 % | 94.26 % | 54.20 % | 90.04 % |
| MeJA-R-3 | 40744494 | 40164434 | 98.36 % | 95.20 % | 54.64 % | 90.73 % |
| MeJA-R-4 | 48265060 | 47530412 | 98.31 % | 95.09 % | 54.41 % | 93.33 % |
Table 2 The quality analysis and mapping statistics of the RNA sequencing data
| 样本 | 原始Reads | 过滤后Reads | Q20 | Q30 | GC含量 | Uniquely mapped |
|---|---|---|---|---|---|---|
| CK-S-1 | 43678100 | 43047426 | 98.30 % | 95.06 % | 55.69 % | 95.74 % |
| CK-S-2 | 50852996 | 50083122 | 98.18 % | 94.75 % | 55.37 % | 95.44 % |
| CK-S-3 | 43444624 | 42661464 | 98.27 % | 94.99 % | 55.15 % | 95.64 % |
| CK-S-4 | 51020748 | 50012420 | 98.19 % | 94.90 % | 56.15 % | 95.19 % |
| CK-R-1 | 47340142 | 46607418 | 98.23 % | 94.92 % | 55.15 % | 93.16 % |
| CK-R-2 | 42318602 | 41636470 | 98.28 % | 95.09 % | 55.11 % | 95.27 % |
| CK-R-3 | 42916752 | 42228274 | 98.32 % | 95.14 % | 54.54 % | 93.34 % |
| CK-R-4 | 47196988 | 46286054 | 98.24 % | 94.94 % | 55.29 % | 93.58 % |
| NaCl-S-1 | 44346810 | 43693600 | 98.41 % | 95.37 % | 55.97 % | 95.90 % |
| NaCl-S-2 | 44482508 | 43736948 | 98.13 % | 94.64 % | 56.02 % | 95.60 % |
| NaCl-S-3 | 45904696 | 45070904 | 98.23 % | 94.89 % | 55.94 % | 96.16 % |
| NaCl-S-4 | 55913340 | 54947042 | 98.20 % | 94.86 % | 55.90 % | 95.91 % |
| NaCl-R-1 | 47132296 | 46300150 | 98.25 % | 95.00 % | 55.83 % | 93.94 % |
| NaCl-R-2 | 42765116 | 42001334 | 98.09 % | 94.64 % | 55.81 % | 93.33 % |
| NaCl-R-3 | 44430198 | 43679556 | 98.17 % | 94.79 % | 56.33 % | 93.88 % |
| NaCl-R-4 | 44579594 | 43831320 | 98.28 % | 95.08 % | 56.02 % | 94.16 % |
| MeJA-S-1 | 53707412 | 52824432 | 98.23 % | 94.92 % | 54.87 % | 94.11 % |
| MeJA-S-2 | 43478130 | 42516802 | 98.24 % | 94.97 % | 54.42 % | 94.95 % |
| MeJA-S-3 | 40559722 | 39809098 | 97.95 % | 94.15 % | 54.50 % | 95.22 % |
| MeJA-S-4 | 46413454 | 45685218 | 98.37 % | 95.28 % | 54.46 % | 95.43 % |
| MeJA-R-1 | 42939854 | 41593670 | 98.30 % | 95.02 % | 54.49 % | 88.28 % |
| MeJA-R-2 | 43131000 | 42474978 | 97.98 % | 94.26 % | 54.20 % | 90.04 % |
| MeJA-R-3 | 40744494 | 40164434 | 98.36 % | 95.20 % | 54.64 % | 90.73 % |
| MeJA-R-4 | 48265060 | 47530412 | 98.31 % | 95.09 % | 54.41 % | 93.33 % |
| 1 | GONG F, YANG L, TAI F, et al.. “Omics” of maize stress response for sustainable food production: opportunities and challenges[J]. OMICS, 2014, 18(12): 714-732. |
| 2 | HICKEY L T, HAFEEZ A N, ROBINSON H, et al.. Breeding crops to feed 10 billion[J]. Nat. Biotechnol., 2019, 37(7): 744-754. |
| 3 | LIU S, ZENDA T, LI J, et al.. Comparative transcriptomic analysis of contrasting hybrid cultivars reveal key drought-responsive genes and metabolic pathways regulating drought stress tolerance in maize at various stages[J/OL]. PLoS One, 2020, 15(10): e0240468[2024-10-24]. . |
| 4 | ZHANG C, YANG R, ZHANG T, et al.. ZmTIFY16, a novel maize TIFY transcription factor gene, promotes root growth and development and enhances drought and salt tolerance in Arabidopsis and Zea mays[J]. Plant Growth Regul., 2023, 100(1): 149-160. |
| 5 | LI C. Breeding crops by design for future agriculture[J]. J. Zhejiang Univ. Sci. B, 2020, 21(6): 423-425. |
| 6 | LIU Y, WANG F, ZHANG A, et al.. Improvement of salinity tolerance in water-saving and drought-resistance rice (WDR)[J/OL]. Int. J. Mol. Sci., 2023, 24(6): 5444[2024-10-24]. . |
| 7 | YANG C, LV D, JIANG S, et al.. Soil salinity regulation of soil microbial carbon metabolic function in the Yellow River Delta, China[J/OL]. Sci. Total Environ., 2021, 790: 148258[2024-10-24]. . |
| 8 | ZHOU J, QIAO J, WANG J, et al.. OsQHB improves salt tolerance by scavenging reactive oxygen species in rice[J/OL]. Front. Plant Sci., 2022, 13: 848891[2024-10-24]. . |
| 9 | ZHAO S, ZHANG Q, LIU M, et al.. Regulation of plant responses to salt stress[J/OL]. Int. J. Mol. Sci., 2021, 22(9): 4609[2024-10-24]. . |
| 10 | GOOSSENS J, FERNÁNDEZ-CALVO P, SCHWEIZER F, et al.. Jasmonates: signal transduction components and their roles in environmental stress responses[J]. Plant Mol. Biol., 2016, 91(6): 673-689. |
| 11 | WANG Y, MOSTAFA S, ZENG W, et al.. Function and mechanism of jasmonic acid in plant responses to abiotic and biotic stresses[J/OL]. Int. J. Mol. Sci., 2021, 22(16): 8568[2024-10-24]. . |
| 12 | WANG J, SONG L, GONG X, et al.. Functions of jasmonic acid in plant regulation and response to abiotic stress[J/OL]. Int. J. Mol. Sci., 2020, 21(4): 1446[2024-10-24]. . |
| 13 | FU J, WU H, MA S, et al.. OsJAZ1 attenuates drought resistance by regulating JA and ABA signaling in rice[J/OL]. Front. Plant Sci., 2017, 8: 2108[2024-10-24]. . |
| 14 | SEO J S, JOO J, KIM M J, et al.. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice[J]. Plant J., 2011, 65(6): 907-921. |
| 15 | DU H, LIU H, XIONG L. Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice[J/OL]. Front. Plant Sci., 2013, 4: 397[2024-10-24]. . |
| 16 | HU Y, JIANG L, WANG F, et al.. Jasmonate regulates the inducer of cbf expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis [J]. Plant Cell, 2013, 25(8): 2907-2924. |
| 17 | WANG L, CHEN H, CHEN G, et al.. Transcription factor SlWRKY50 enhances cold tolerance in tomato by activating the jasmonic acid signaling[J]. Plant Physiol., 2024, 194(2): 1075-1090. |
| 18 | DING F, WANG C, ZHANG S, et al.. A jasmonate-responsive glutathione S-transferase gene SlGSTU24 mitigates cold-induced oxidative stress in tomato plants[J/OL]. Sci. Hortic., 2022, 303: 111231[2024-10-24]. . |
| 19 | SHANG C, LIU X, CHEN G, et al.. SlWRKY81 regulates Spd synthesis and Na(+)/K(+) homeostasis through interaction with SlJAZ1 mediated JA pathway to improve tomato saline-alkali resistance[J]. Plant J., 2024, 118(6): 1774-1792. |
| 20 | WU H, YE H, YAO R, et al.. OsJAZ9 acts as a transcriptional regulator in jasmonate signaling and modulates salt stress tolerance in rice[J]. Plant Sci., 2015, 232: 1-12. |
| 21 | QIU Z, GUO J, ZHU A, et al.. Exogenous jasmonic acid can enhance tolerance of wheat seedlings to salt stress[J]. Ecotoxicol. Environ. Saf., 2014, 104: 202-208. |
| 22 | 王芳, 周娟,黄兴华, 等. 外源MeJA对盐胁迫下玉米幼苗生长及抗氧化酶基因表达的影响[J]. 玉米科学, 2022, 30(2): 75-81. |
| WANG F, ZHOU J, HUANG X H, et al.. Effects of exogenous MeJA on growth and antioxidant enzyme gene expression of maize seedlings under salt stress[J]. J. Maize Sci., 2022, 30(2): 75-81. | |
| 23 | 陈芳, 杨双龙, 张莉, 等. 外源茉莉酸甲酯对盐胁迫下玉米幼苗AsA-GSH循环的影响[J]. 生物学通报, 2021, 56(11): 44-48. |
| CHEN F, YANG S L, ZHANG L, et al.. Effects of exogenous methyl jasmonate on ascorbate-glutathione cycle in Zea mays seedlings under salt stress[J]. Bull. Biol., 2021, 56(11): 44-48. | |
| 24 | YU Z, DUAN X, LUO L, et al.. How plant hormones mediate salt stress responses[J]. Trends Plant Sci., 2020, 25(11): 1117-1130. |
| 25 | LIU S, ZHANG P, LI C, et al.. The moss jasmonate ZIM-domain protein PnJAZ1 confers salinity tolerance via crosstalk with the abscisic acid signalling pathway[J]. Plant Sci., 2019, 280: 1-11. |
| 26 | 史庆玲, 李忠峰, 董永彬, 等. 植物乙烯信号转导通路及其相关基因的研究进展[J]. 生物技术进展, 2019, 9(5): 449-454. |
| SHI Q L, LI Z F, DONG Y B, et al.. Progress on ethylene signal transduction pathway and related genes in plants[J]. Curr. Biotechnol., 2019, 9(5): 449-454. | |
| 27 | RAZA A, CHARAGH S, ZAHID Z, et al.. Jasmonic acid: a key frontier in conferring abiotic stress tolerance in plants[J]. Plant Cell Rep., 2021, 40(8): 1513-1541. |
| 28 | CAO H, ZHANG K, LI W, et al.. ZmMYC7 directly regulates ZmERF147 to increase maize resistance to Fusarium graminearum [J]. Crop J., 2023, 11(1): 79-88. |
| 29 | MA C, LI R, SUN Y, et al.. ZmMYC2s play important roles in maize responses to simulated herbivory and jasmonate[J]. J. Integr. Plant Biol., 2023, 65(4): 1041-1058. |
| 30 | HE Y, BORREGO E J, GORMAN Z, et al.. Relative contribution of LOX10, green leaf volatiles and JA to wound-induced local and systemic oxylipin and hormone signature in Zea mays (maize)[J/OL]. Phytochemistry, 2020, 174: 112334[2024-10-24]. . |
| 31 | ZHANG X, LIU P, QING C, et al.. Comparative transcriptome analyses of maize seedling root responses to salt stress[J/OL]. PeerJ, 2021, 9: e10765[2024-10-24]. . |
| 32 | KAZAN K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance[J]. Trends Plant Sci., 2015, 20(4): 219-229. |
| 33 | WANG M, FAN X, DING F. Jasmonate: a hormone of primary importance for temperature stress response in plants[J/OL]. Plants (Basel), 2023, 12(24): 4080[2024-10-24]. . |
| 34 | WANG W, CAO J, HUANG S, et al.. Integrated transcriptomics and metabolomics analyses provide insights into salt-stress response in germination and seedling stage of wheat (Triticum aestivum L.)[J/OL]. Curr. Plant Biol., 2023, 33: 100274[2024-10-24]. . |
| 35 | XIONG H, GUO H, XIE Y, et al.. RNAseq analysis reveals pathways and candidate genes associated with salinity tolerance in a spaceflight-induced wheat mutant[J/OL]. Sci. Rep., 2017, 7(1): 2731[2024-10-24]. . |
| 36 | LUO Q, TENG W, FANG S, et al.. Transcriptome analysis of salt-stress response in three seedling tissues of common wheat[J]. Crop J., 2019, 7(3): 378-392. |
| 37 | XIONG Y, YAN H, LIANG H, et al.. RNA-Seq analysis of Clerodendrum inerme (L.) roots in response to salt stress[J/OL]. BMC Genomics, 2019, 20(1): 724[2024-10-24]. . |
| 38 | ZHU M, LIU Y, CAI P, et al.. Jasmonic acid pretreatment improves salt tolerance of wheat by regulating hormones biosynthesis and antioxidant capacity[J/OL]. Front. Plant Sci., 2022, 13: 968477[2024-10-24]. . |
| 39 | LIU S, HE Y, FU Y, et al.. Transcriptome sequencing revealed the mechanism of promoting floret opening by exogenous methyl jasmonate in Sorghum [J/OL]. 3 Biotech, 2021, 11(4): 181[2024-10-24]. . |
| 40 | NIE G, ZHOU J, JIANG Y, et al.. Transcriptome characterization of candidate genes for heat tolerance in perennial ryegrass after exogenous methyl Jasmonate application[J/OL]. BMC Plant Biol., 2022, 22(1): 68[2024-10-24]. . |
| 41 | ZHU J, WEI X, YIN C, et al.. ZmEREB57 regulates OPDA synthesis and enhances salt stress tolerance through two distinct signalling pathways in Zea mays [J]. Plant Cell Environ., 2023, 46(9): 2867-2883. |
| 42 | JIANG J, MA S, YE N, et al.. WRKY transcription factors in plant responses to stresses[J]. J. Integr. Plant Biol., 2017, 59(2): 86-101. |
| 43 | LUO P, CHEN Y, RONG K, et al.. ZmSNAC13, a maize NAC transcription factor conferring enhanced resistance to multiple abiotic stresses in transgenic Arabidopsis [J]. Plant Physiol. Biochem., 2022, 170: 160-170. |
| 44 | WU J, JIANG Y, LIANG Y, et al.. Expression of the maize MYB transcription factor ZmMYB3R enhances drought and salt stress tolerance in transgenic plants[J]. Plant Physiol. Biochem., 2019, 137: 179-188. |
| 45 | CHENG C, AN L, LI F, et al.. Wide-range portrayal of AP2/ERF transcription factor family in maize (Zea mays L.) development and stress responses[J/OL]. Genes (Basel), 2023, 14(1): 194[2024-10-24]. . |
| 46 | WANG Y M, YANG Q, XU H, et al.. Physiological and transcriptomic analysis provide novel insight into cobalt stress responses in willow[J/OL]. Sci. Rep., 2020, 10(1): 2308[2024-10-24]. . |
| 47 | MATHAN J, SINGH A, RANJAN A. Sucrose transport in response to drought and salt stress involves ABA-mediated induction of OsSWEET13 and OsSWEET15 in rice[J]. Physiol. Plant., 2021, 171(4): 620-637. |
| 48 | ZHANG J, ZHOU Y, LIU L, et al.. Overexpression of OsSWEET5 in rice causes growth retardation and precocious senescence[J/OL]. PLoS One, 2014, 9(4): e94210[2024-10-24]. . |
| 49 | GAUTAM T, DUTTA M, JAISWAL V, et al.. Emerging roles of SWEET sugar transporters in plant development and abiotic stress responses[J/OL]. Cells, 2022, 11(8): 1303[2024-10-24]. . |
| 50 | ROBERT C A M, MATEO P. The chemical ecology of benzoxazinoids[J]. Chimia (Aarau), 2022, 76(11): 928-938. |
| 51 | 王颖, 梅馨月, 刘屹湘, 等. 植物中防御相关物质苯并嗯嗪类的研究进展[J]. 植物生理学报, 2021, 57(4): 767-779. |
| WANG Y, MEI X Y, LIU Y X, et al.. Research progress of benzoxazinoids as defense related substances in plants[J]. Plant Physiol. J., 2021, 57(4): 767-779. |
| [1] | Lifeng E, Yani LI, Yu XIE, Jianhua QUAN, Xiubin CHEN, Jun HUA, Xuemei ZHAO, Wenqin ZHAO. Physiological and Photosynthetic Responses of Brassica pekinensis Seedlings to Salt Stress Alleviation by Chitosan Oligosaccharide [J]. Current Biotechnology, 2025, 15(4): 675-682. |
| [2] | AYELHAN·Haysa, Fei JIAO, Hong LIU, ANASI·Hudelati, Yubang SHEN. Integrating GWAS and Transcriptome Profiling to Identify SNP Markers Linked to High-temperature Tolerance in Esox lucius [J]. Current Biotechnology, 2025, 15(3): 432-445. |
| [3] | Qingyun ZHANG, Lei MA, Hua XU, Lian JIN, Junjie ZOU, Baobao WANG, Quanjia CHEN, Miaoyun XU. Mechanism of Lignin Content in Root System Affecting Salt Tolerance in Maize [J]. Current Biotechnology, 2025, 15(1): 67-77. |
| [4] | Xin QI, Xinran LI, Yaning GUO, Dan WANG, Kai LI, Qiong WU, Liang LI. Comparison of Endogenous Genes in Maize Based on Digital PCR [J]. Current Biotechnology, 2025, 15(1): 78-85. |
| [5] | Jingsheng TAN, Guantao ZHAO, Li ZHU, Yubin LI. The Sequence Characterization of Somatic Excision Footprints from Two-element Transposable System of rMrh/Mrh in Maize [J]. Current Biotechnology, 2024, 14(4): 601-609. |
| [6] | Yezi MA, Meijuan XIA, Cuicui LIU, Hongtao WANG, Jiaxi ZHOU. Comparative Analysis of Platelet Transcriptome and Proteome Changes in SARS-CoV2 Omicron Infection [J]. Current Biotechnology, 2024, 14(4): 649-656. |
| [7] | Qiyu ZHANG, Xiaogui LYU, Jin BAO. Effect of Cold Plasma Treatment on Seed Germination and Seedling Growth of Oats Under Salt Stress [J]. Current Biotechnology, 2024, 14(3): 413-421. |
| [8] | Hui ZHANG, Bobo LIU, Fengmei LI, Jian CUI. Study on the Involvement of Auxin Signaling Pathway in Response to Powdery Mildew Stress in Pumpkin [J]. Current Biotechnology, 2024, 14(3): 433-441. |
| [9] | Hongmei QIAO. Transcriptome Analysis of the Terrestrial Plant Carallia brachiata from Rhizophoraceae [J]. Current Biotechnology, 2024, 14(2): 228-236. |
| [10] | Guantao ZHAO, Jingsheng TAN, Li ZHU, Yubin LI. The Sequence Characterization of the Somatic Footprints upon the Transposition of Non-autonomous Transposon rDt in Maize [J]. Current Biotechnology, 2024, 14(2): 248-256. |
| [11] | Lingyan LI, Bing XIAO, Xudong ZHANG, Hua ZHANG, Ziyan CHEN, Haoqian WANG, Xiujie ZHANG, Hong CHEN, Jingang LIANG. Establishment and Standardization of Real-time PCR Method for Qualitative Detection of Genetically Modified Maize MON87411 [J]. Current Biotechnology, 2024, 14(2): 257-262. |
| [12] | Caihong WANG, Yuhan SHAO, Mengyuan JIANG. The Mechanisms of IL-17 Inhibiting Pseudomonas aeruginosa Infection on Lung Epithelial Cells [J]. Current Biotechnology, 2024, 14(2): 304-311. |
| [13] | Qinqin LIU, Baiming HUANG, Yezi MA, Cuicui LIU, Hongtao WANG, Jiaxi ZHOU. Comparative Analysis of Megakaryocyte and Platelet Transcriptome Changes in Acute Infection [J]. Current Biotechnology, 2023, 13(3): 465-472. |
| [14] | Min LI, Lei WANG, Junjie ZOU. Opportunities and Challenges for the Industrial Application of Transgenic Insect-resistant and Herbicide-tolerant Maize in China [J]. Current Biotechnology, 2023, 13(2): 157-165. |
| [15] | Ke XIAO, Xiaojin ZHOU, Rumei CHEN, Sen PANG. Interactions Between ClassⅠand ClassⅡ NAS Proteins in Maize Using Bimolecular Fluorescence Complementation (BiFC) Assay [J]. Current Biotechnology, 2022, 12(5): 728-736. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||