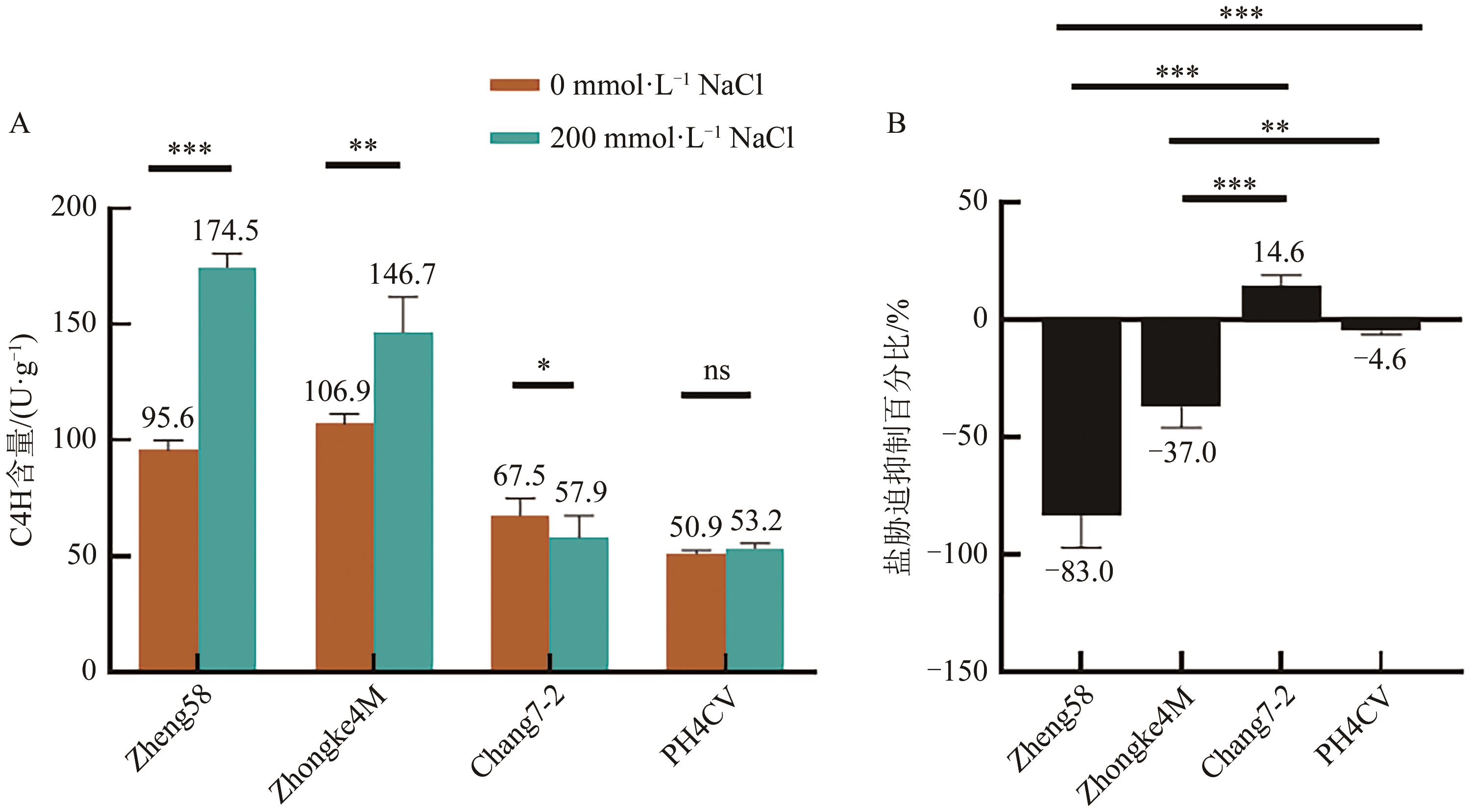
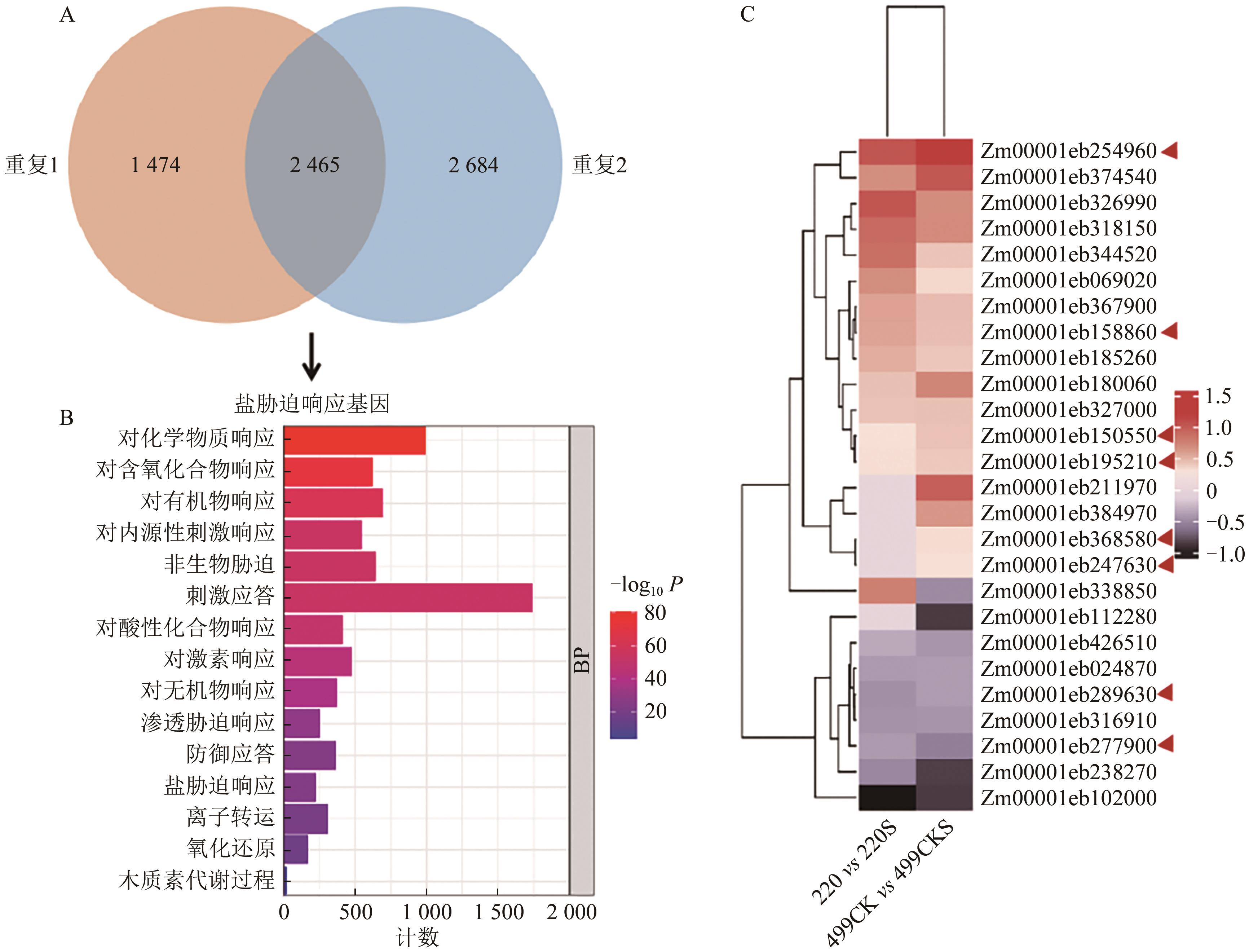
| 1 |
YANG Y, GUO Y. Elucidating the molecular mechanisms mediating plant salt-stress responses[J]. New Phytol., 2018, 217(2): 523-539.
|
| 2 |
阿提开姆·麦麦提,顾炜,于典司,等.基于隶属函数法的玉米种质资源苗期耐盐性评价[J].上海农业学报,2023,39(5):54-60.
|
| 3 |
王继伟,赵成章,赵连春,等.内陆盐沼芦苇根系形态及生物量分配对土壤盐分因子的响应[J].生态学报,2018,38(13):4843-4851.
|
|
WANG J W, ZHAO C Z, ZHAO L C, et al.. Response of root morphology and biomass of Phragmites australis to soil salinity in inland salt marsh[J]. Acta Ecol. Sin., 2018, 38(13): 4843-4851.
|
| 4 |
BYRT C S, MUNNS R, BURTON R A, et al.. Root cell wall solutions for crop plants in saline soils[J]. Plant Sci., 2018, 269: 47-55.
|
| 5 |
CHOAT B, DRAYTON W M, BRODERSEN C, et al.. Measurement of vulnerability to water stress-induced cavitation in grapevine: a comparison of four techniques applied to a long-vesseled species[J]. Plant Cell Environ., 2010, 33(9): 1502-1512.
|
| 6 |
SELLAMI S, LE HIR R, THORPE M R, et al.. Arabidopsis natural accessions display adaptations in inflorescence growth and vascular anatomy to withstand high salinity during reproductive growth[J/OL]. Plants (basel switz.), 2019, 8(3): E61[2024-10-15]. .
|
| 7 |
HORIE T, KARAHARA I, KATSUHARA M. Salinity tolerance mechanisms in glycophytes: an overview with the central focus on rice plants[J/OL]. Rice, 2012, 5(1): 11[2024-10-15]..
|
| 8 |
YU J, ZAO W G, HE Q, et al.. Genome-wide association study and gene set analysis for understanding candidate genes involved in salt tolerance at the rice seedling stage[J]. Mol. Genet. Genom., 2017, 292(6): 1391-1403.
|
| 9 |
ANDERSON C T, KIEBER J J. Dynamic construction, perception, and remodeling of plant cell walls[J]. Annu. Rev. Plant Biol., 2020, 71: 39-69.
|
| 10 |
LIU J, ZHANG W, LONG S, et al.. Maintenance of cell wall integrity under high salinity[J/OL]. Int. J. Mol. Sci., 2021, 22(6): 3260[2024-10-15]. .
|
| 11 |
DABRAVOLSKI S A, ISAYENKOV S V. The regulation of plant cell wall organisation under salt stress[J/OL]. Front. Plant Sci., 2023, 14: 1118313[2024-10-15]. .
|
| 12 |
ZHANG B, GAO Y, ZHANG L, et al.. The plant cell wall: Biosynthesis, construction,and functions[J]. J. Integr. Plant Biol., 2021, 63(1):251-272.
|
| 13 |
LI H, YAN S H, ZHAO L, et al.. Histone acetylation associated up-regulation of the cell wall related genes is involved in salt stress induced maize root swelling[J/OL]. BMC Plant Biol., 2014, 14(1): 105[2024-11-15]. .
|
| 14 |
KESTEN C, GARCÍA-MORENO Á, AMORIM-SILVA V, et al.. Peripheral membrane proteins modulate stress tolerance by safeguarding cellulose synthases[J/OL]. Sci. Adv., 2022, 8(46): eabq6971[2024-10-15]. .
|
| 15 |
NAGASHIMA Y, MA Z, LIU X, et al.. Multiple quality control mechanisms in the ER and TGN determine subcellular dynamics and salt-stress tolerance function of KORRIGAN1[J]. Plant cell, 2020, 32(2): 470-485.
|
| 16 |
DEGENHARDT B, GIMMLER H. Cell wall adaptations to multiple environmental stresses in maize roots[J]. Arthritis Res.Ther., 2000, 51(344): 595-603.
|
| 17 |
QIN R D, HU Y M, CHEN H, et al.. MicroRNA408 negatively regulates salt tolerance by affecting secondary cell wall development in maize[J]. Plant Physiol., 2023, 192(2): 1569-1583.
|
| 18 |
VANHOLME R, DE MEESTER B, RALPH J, et al.. Lignin biosynthesis and its integration into metabolism[J]. Curr. Opin. Biotechnol., 2019, 56: 230-239.
|
| 19 |
GUO H, WANG Y, WANG L, et al.. Expression of the MYB transcription factor gene BplMYB46 affects abiotic stress tolerance and secondary cell wall deposition in Betula platyphylla [J]. Plant Biotechnol. J., 2017, 15(1): 107-121.
|
| 20 |
KONG Q S, MOSTAFA H H A, YANG W L, et al.. Comparative transcriptome profiling reveals that brassinosteroid-mediated lignification plays an important role in garlic adaption to salt stress[J]. Plant Physiol. Biochem., 2021, 158: 34-42.
|
| 21 |
WANG B B, LIN Z C, LI X, et al.. Genome-wide selection and genetic improvement during modern maize breeding[J]. Nat. Genet., 2020, 52: 565-571.
|
| 22 |
SILVA B R S, BATISTA B L, LOBATO A K S. Anatomical changes in stem and root of soybean plants submitted to salt stress[J]. Plant Biol. (stuttgart ger.), 2021, 23(1): 57-65.
|
| 23 |
ROGERS L A, DUBOS C, SURMAN C, et al.. Comparison of lignin deposition in three ectopic lignification mutants[J]. New Phytol., 2005, 168(1): 123-140.
|
| 24 |
VAN ZELM E, ZHANG Y, TESTERINK C. Salt tolerance mechanisms of plants[J]. Annu. Rev. Plant Biol., 2020, 71: 403-433.
|
 ), Lei MA2, Hua XU2, Lian JIN2, Junjie ZOU2,3, Baobao WANG2,3, Quanjia CHEN1(
), Lei MA2, Hua XU2, Lian JIN2, Junjie ZOU2,3, Baobao WANG2,3, Quanjia CHEN1( ), Miaoyun XU2,3(
), Miaoyun XU2,3( )
)
 ), 马蕾2, 胥华2, 靳礼安2, 邹俊杰2,3, 王宝宝2,3, 陈全家1(
), 马蕾2, 胥华2, 靳礼安2, 邹俊杰2,3, 王宝宝2,3, 陈全家1( ), 徐妙云2,3(
), 徐妙云2,3( )
)