Current Biotechnology ›› 2022, Vol. 12 ›› Issue (2): 168-175.DOI: 10.19586/j.2095-2341.2021.0140
• Reviews • Previous Articles Next Articles
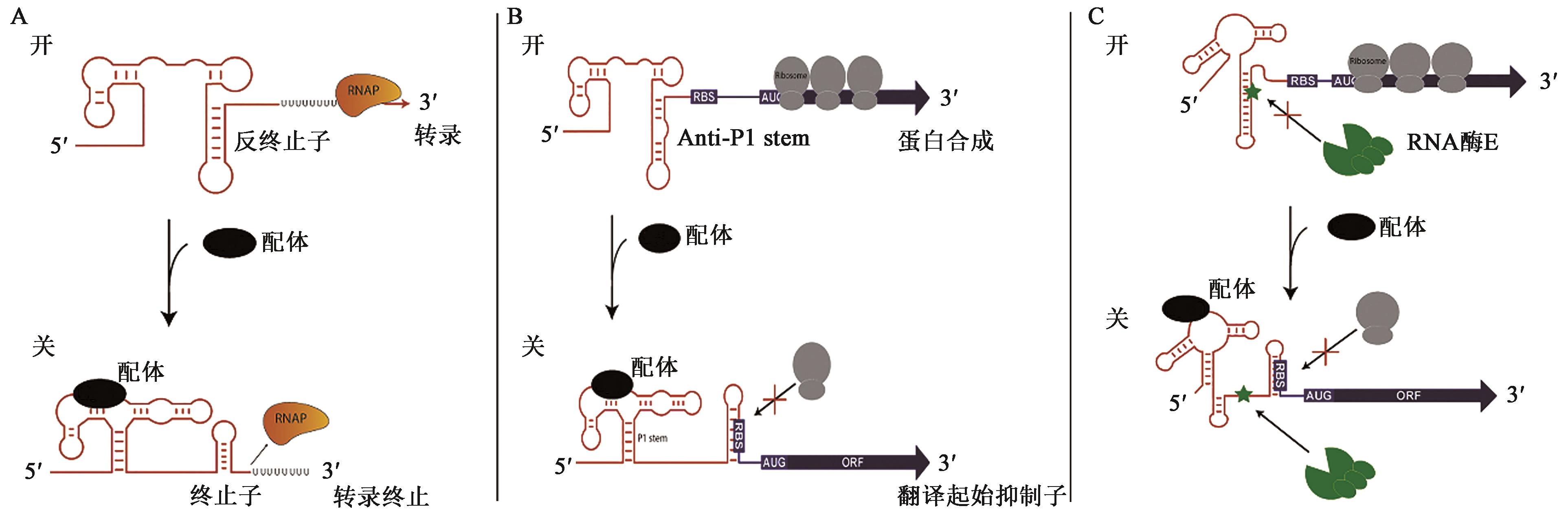
Research Progress of Riboswitch Biosensors for Small Molecule Target
Yifan WU1( ), Shenghao LIN2, Wentao XU1,2(
), Shenghao LIN2, Wentao XU1,2( )
)
- 1.College of Food Science and Nutritional Engineering,China Agricultural University,Beijing 100083,China
2.Key Laboratory of Precision Nutrition and Food Quality,Department of Nutrition and Health,China Agricultural University,Beijing 100083,China
-
Received:2021-08-05Accepted:2021-09-03Online:2022-03-25Published:2022-03-25 -
Contact:Wentao XU
小分子靶标的核糖开关生物传感器研究进展
- 1.中国农业大学食品科学与营养工程学院,北京 100083
2.中国农业大学营养与健康系,食品精准营养与质量控制教育部重点实验室,北京 100083
-
通讯作者:许文涛 -
作者简介:吴一凡 E-mail: Wyf001208@126.com; -
基金资助:山东省重大科技创新工程项目(2019JZZY011014);北京市农业科技项目(PXM2021_014207_000037)
CLC Number:
Cite this article
Yifan WU, Shenghao LIN, Wentao XU. Research Progress of Riboswitch Biosensors for Small Molecule Target[J]. Current Biotechnology, 2022, 12(2): 168-175.
吴一凡, 林晟豪, 许文涛. 小分子靶标的核糖开关生物传感器研究进展[J]. 生物技术进展, 2022, 12(2): 168-175.
share this article
| 传感器类别 | 优点 | 缺点 |
|---|---|---|
| 核糖开关传感器 | 应用范围广,识别元件可快速响应,高特异性和灵敏度,成本低 | 结构刚性不足,人工合成筛选 |
| 酶生物传感器 | 制备简便,高特异性 | 应用范围相对较窄,成本高,易失活,稳定性较低 |
| 免疫传感器 | 抗原抗体识别特异性好 | 应用范围相对较窄,成本高,稳定性较低 |
| 微生物传感器 | 成本低,耐久性好 | 稳定性、灵敏度低,响应时间久 |
| 细胞传感器 | 成本低,制作简便 | 应用范围有限,稳定性较低 |
Table 1 The characteristics of different types of biosensors
| 传感器类别 | 优点 | 缺点 |
|---|---|---|
| 核糖开关传感器 | 应用范围广,识别元件可快速响应,高特异性和灵敏度,成本低 | 结构刚性不足,人工合成筛选 |
| 酶生物传感器 | 制备简便,高特异性 | 应用范围相对较窄,成本高,易失活,稳定性较低 |
| 免疫传感器 | 抗原抗体识别特异性好 | 应用范围相对较窄,成本高,稳定性较低 |
| 微生物传感器 | 成本低,耐久性好 | 稳定性、灵敏度低,响应时间久 |
| 细胞传感器 | 成本低,制作简便 | 应用范围有限,稳定性较低 |
| 1 | 胡鹏. 核酸适体在蛋白质和小分子检测中的新方法研究[D]. 湖南:湖南大学, 2010. |
| 2 | MACHTEL P, BĄKOWSKA Ż K, ŻYWICKI M. Emerging applications of riboswitches——from antibacterial targets to molecular tools[J]. J. Appl. Genet., 2016,57(4): 531-541. |
| 3 | TURNER A P F. Biosensors: sense and sensibility[J]. Chem. Soc. Rev., 2013,42(8): 3184-3196. |
| 4 | 张泽, 张颖聪, 于洪伟, 等. 生物传感器识别元件的种类及其在临床检验中的研究进展[J]. 临床检验杂志, 2020,38(10): 767-771. |
| 5 | YOKOBAYASHI Y. Aptamer-based and aptazyme-based riboswitches in mammalian cells[J]. Curr. Opin. Chem. Biol., 2019,52: 72-78. |
| 6 | BLOUIN S, MULHBACHER J, PENEDO J C, et al.. Riboswitches: ancient and promising genetic regulators[J]. ChemBioChem, 2009,10(3): 400-416. |
| 7 | BAIRD N J, KULSHINA N, FERRE-D'AMARE A R. Riboswitch function flipping the switch or tuning the dimmer?[J]. RNA Biol., 2010,7(3): 328-332. |
| 8 | BREAKER R R. Riboswitches and the RNA World[J/OL]. Cold Spring Harbor Perspect. Biol., 2012,4(2):a3566[2022-02-20]. . |
| 9 | GOLD L, BROWN D, HE Y Y, et al.. From oligonucleotide shapes to genomic SELEX: novel biological regulatory loops[J]. Proc. Natl. Acad. Sci. USA, 1997,94(1): 59-64. |
| 10 | GOLD L, SINGER B, HE Y Y, et al.. SELEX and the evolution of genomes[J]. Curr. Opin. Genet. Devlop., 1997,7(6): 848-851. |
| 11 | GELFAND M S, MIRONOV A A, JOMANTAS J, et al.. A conserved RNA structure element involved in the regulation of bacterial riboflavin synthesis genes[J]. Trends Genet., 1999,15(11): 439-442. |
| 12 | WINKLER W, NAHVI A, BREAKER R R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression[J]. Nature, 2002,419(6910): 952-956. |
| 13 | MCCOWN P J, CORBINO K A, STAV S, et al.. Riboswitch diversity and distribution[J]. RNA, 2017,23(7): 995-1011. |
| 14 | SUBBAIAH K C V, HEDAYA O, WU J, et al.. Mammalian RNA switches: molecular rheostats in gene regulation, disease, and medicine[J]. Comput. Struct. Biotechnol. J., 2019,17: 1326-1338. |
| 15 | MACHTEL P, BAKOWSKA-ZYWICKA K, ZYWICKI M. Emerging applications of riboswitches - from antibacterial targets to molecular tools[J]. J. Appl. Genet., 2016,57(4): 531-541. |
| 16 | SCULL C E, DANDPAT S S, ROMERO R A, et al.. Transcriptional riboswitches integrate timescales for bacterial gene expression control[J/OL]. Front. Mol. Biosci., 2021,7(607158):607158[2022-02-20]. . |
| 17 | 熊莹喆, 曹苑青, 肖玲慧, 等. 基于核糖开关的新型基因表达调控系统的应用[J]. 生物技术通报, 2017,33(2): 41-46. |
| 18 | BÉDARD A V, HIEN E D M, LAFONTAINE D A. Riboswitch regulation mechanisms: RNA, metabolites and regulatory proteins[J/OL]. Biochim. Biophys. Acta Gene Regul. Mechan., 2020,1863(3): 194501[2022-02-20]. . |
| 19 | BARRICK J E, BREAKER R R. The distributions, mechanisms, and structures of metabolite-binding riboswitches[J/OL]. Genome Biol., 2007,8(11): R239[2022-02-20]. . |
| 20 | DOMIN G, FINDEISS S, WACHSMUTH M, et al.. Applicability of a computational design approach for synthetic riboswitches[J]. Nucl. Acids Res., 2017,45(7): 4108-4119. |
| 21 | BORUJENI A E, MISHLER D M, WANG J, et al.. Automated physics-based design of synthetic riboswitches from diverse RNA aptamers[J]. Nucl. Acids Res., 2016,44(1): 1-13. |
| 22 | ELLINGTON A D, SZOSTAK J W. In vitro selection of RNA molecules that bind specific ligands[J]. Nature, 1990,346(6287): 818-822. |
| 23 | WITTMANN A, SUESS B. Engineered riboswitches: expanding researchers' toolbox with synthetic RNA regulators[J]. FEBS Lett., 2012,586(15): 2076-2083. |
| 24 | MURATA A, SATO S. In vitro selection of RNA aptamers for a small-molecule dye[M]//OGAWA A. Methods in molecular biology, 2014: 17-28. |
| 25 | FINDEISS S, ETZEL M, WILL S, et al.. Design of artificial riboswitches as viosensors[J/OL]. Sensors, 2017,17(19909):1990[2022-02-20]. . |
| 26 | REYNOSO C M K, MILLER M A, BINA J E, et al.. Riboswitches for intracellular study of genes involved in Francisella pathogenesis[J/OL]. mBIO, 2012,3(6):e00253-12[2022-02-20]. . |
| 27 | WACHSMUTH M, FINDEISS S, WEISSHEIMER N, et al.. De novo design of a synthetic riboswitch that regulates transcription termination[J]. Nucl. Acids Res., 2013,41(4): 2541-2551. |
| 28 | HANSON S, BAUER G, FINK B, et al.. Molecular analysis of a synthetic tetracycline-binding riboswitch[J]. RNA, 2005,11(4): 503-511. |
| 29 | XIAO H, EDWARDS T E, FERRE-D'AMARE A R. Structural vasis for specific, high-affinity tetracycline binding by an in vitro evolved aptamer and artificial riboswitch[J]. Chem. Biol., 2008,15(10): 1125-1137. |
| 30 | WEIGAND J E, SUESS B. Tetracycline aptamer-controlled regulation of pre-mRNA splicing in yeast[J]. Nucl. Acids Res., 2007,35(12): 4179-4185. |
| 31 | LINK K H, BREAKER R R. Engineering ligand-responsive gene-control elements: lessons learned from natural riboswitches[J]. Gene Ther., 2009,16(10): 1189-1201. |
| 32 | SINHA J, REYES S, GALLIVAN J P. Reprogramming bacteria to seek and destroy an herbicide [J/OL]. Nat. Chem. Biol., 2014,10(3): 239[2022-02-20].. |
| 33 | WEIGAND J E, SANCHEZ M, GUNNESCH E, et al.. Screening for engineered neomycin riboswitches that control translation initiation[J]. RNA, 2008,14(1): 89-97. |
| 34 | 杨会勇, 刁勇, 林俊生, 等. 新型基因表达调控元件——人工核糖开关的构建及筛选[J]. 生物工程学报, 2012,28(2): 134-143. |
| 35 | NOMURA Y, YOKOBAYASHI Y. Reengineering a natural riboswitch by dual genetic selection[J/OL]. J. Am. Chem. Soc., 2007,129(45): 13814[2022-02-20]. . |
| 36 | LYNCH S A, DESAI S K, SAJJA H K, et al.. A high-throughput screen for synthetic riboswitches reveals mechanistic insights into their function[J]. Chem. Biol., 2007,14(2): 173-184. |
| 37 | KIRCHNER M, SCHORPP K, HADIAN K, et al.. An in vivo high-throughput screening for riboswitch ligands using a reverse reporter gene system[J/OL]. Sci. Rep., 2017,7:7732[2021-08-28]. . |
| 38 | PARKHEY P, MOHAN S V. Biosensing applications of microbial fuel cell: approach toward miniaturization[M]//MOHAN S V, VARJANI S, PANDEY A. Biomass biofuels biochemicals. 2019:977-997. |
| 39 | 石亚丽, 袁涛, 任婷婷, 等. 生物传感器在食品安全快速检测中应用研究[J]. 粮食与油脂, 2012,25(2): 5-9. |
| 40 | SERGANOV A, NUDLER E. A decade of riboswitches[J]. Cell, 2013,152(1-2): 17-24. |
| 41 | FOWLER C C, BROWN E D, LI Y. A FACS-based approach to engineering artificial riboswitches[J]. ChemBioChem, 2008,9(12): 1906-1911. |
| 42 | YOU M, LITKE J L, JAFFREY S R. Imaging metabolite dynamics in living cells using a spinach-based riboswitch[J]. Proc. Natl. Acad. Sci. USA, 2015,112(21): E2756-E2765. |
| 43 | KELLENBERGER C A, HAMMOND M C. In vitro analysis of riboswitch-spinach aptamer fusions as metabolite-sensing fluorescent biosensors[M]//BURKEAGUERO D H. Methods in Enzymology. 2015:147-172. |
| 44 | KELLENBERGER C A, WILSON S C, SALES-LEE J, et al.. RNA-based fluorescent biosensors for live cell imaging of second messengers cyclic di-GMP and cyclic AMP-GMP[J]. J. Am. Chem. Soc., 2013,135(13): 4906-4909. |
| 45 | DASGUPTA S, SHELKE S A, LI N, et al.. Spinach RNA aptamer detects lead (Ⅱ) with high selectivity[J]. Chem. Commun., 2015,51(43): 9034-9037. |
| 46 | SAVAGE J C, SHINDE P, BACHINGER H P, et al.. A ribose modification of Spinach aptamer accelerates lead(ii) cation association in vitro[J]. Chem. Commun., 2019,55(42): 5882-5885. |
| 47 | FILONOV G S, MOON J D, SVENSEN N, et al.. Broccoli: rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution[J]. J. Am. Chem. Soc., 2014,136(46): 16299-16308. |
| 48 | DOLGOSHEINA E V, JENG S C Y, PANCHAPAKESAN S S S, et al.. RNA mango aptamer-fluorophore: a bright, high-affinity complex for RNA labeling and tracking[J]. ACS Chem. Biol., 2014,9(10): 2412-2420. |
| 49 | MURANAKA N, SHARMA V, NOMURA Y, et al.. An efficient platform for genetic selection and screening of gene switches in Escherichia coli [J/OL]. Nucl. Acids. Res., 2009,37(5):e39 [2021-08-28]. . |
| 50 | DONOVAN P D, HOLLAND L M, LOMBARDI L, et al.. TPP riboswitch-dependent regulation of an ancient thiamin transporter in Candida[J/OL]. PLoS Genet., 2018,14(5): e1007429[2022-02-20]. . |
| 51 | MOLDOVAN M A, PETROVA S A, GELFAND M S. Comparative genomic analysis of fungal TPP-riboswitches[J]. Fungal Genet. Biol., 2018,114: 34-41. |
| 52 | CROFT M T, MOULIN M, WEBB M E, et al.. Thiamine biosynthesis in algae is regulated by riboswitches[J]. Proc. Natl. Acad. Sci. USA, 2007,104(52): 20770-20775. |
| 53 | SUBKI A, HO C L, ISMAIL N, et al.. Identification and characterisation of thiamine pyrophosphate (TPP) riboswitch in Elaeis guineensis[J/OL]. PLoS ONE, 2020,15(7): e235431[2022-02-20].. |
| 54 | BASTET L, TURCOTTE P, WADE J T, et al.. Maestro of regulation: riboswitches orchestrate gene expression at the levels of translation, transcription and mRNA decay[J]. RNA Biol., 2018,15(6): 679-682. |
| 55 | AGHDAM E M, SINN M, TARHRIZ V, et al.. TPP riboswitch characterization in Alishewanella tabrizica and Alishewanella aestuarii and comparison with other TPP riboswitches[J]. Microbiol. Res., 2017,195: 71-80. |
| 56 | PAVLOVA N, PENCHOVSKY R. Genome-wide bioinformatics analysis of FMN, SAM-I, glmS, TPP, lysine, purine, cobalamin, and SAH riboswitches for their applications as allosteric antibacterial drug targets in human pathogenic bacteria[J]. Exp. Opin. Therap. Targets, 2019,23(7): 631-643. |
| 57 | PRICE I R, GRIGG J C, KE A. Common themes and differences in SAM recognition among SAM riboswitches[J]. Biochim. Biophys. Acta Gene Regul. Mechan., 2014,1839(10SI): 931-938. |
| 58 | TANG D, DU X, SHI Q, et al.. A SAM-I riboswitch with the ability to sense and respond to uncharged initiator tRNA[J/OL]. Nat. Commun., 2020,11(1):2794 [2021-08-28]. . |
| 59 | ST-PIERRE P, SHAW E, JACQUES S, et al.. A structural intermediate pre-organizes the add adenine riboswitch for ligand recognition[J]. Nucl. Acids Res., 2021,49(10): 5891-5904. |
| 60 | HU G, LI H, XU S, et al.. Ligand binding mechanism and its relationship with conformational changes in adenine riboswitch[J/OL]. Intern. J. Mol. Sci., 2020,21(6):1926[2022-02-20]. . |
| 61 | 张蕾. 腺嘌呤核糖开关的去折叠路径的多尺度模拟[D]. 北京:北京工业大学, 2015. |
| 62 | 凌宝萍. 嘌呤核糖开关和细胞凋亡抑制蛋白与药物作用机制的理论研究[D]. 山东:山东大学, 2010. |
| 63 | LEMAY J, DESNOYERS G, BLOUIN S, et al.. Comparative study between transcriptionally-and translationally-acting adenine riboswitches reveals key differences in riboswitch regulatory mechanisms[J/OL]. PLoS Genet., 2011,7(1):e1001278 [2021-08-28]. . |
| 64 | PAVLOVA N, KALOUDAS D, PENCHOVSKY R. Riboswitch distribution, structure, and function in bacteria[J]. Gene, 2019,708: 38-48. |
| 65 | SERGANOV A, YUAN Y R, PIKOVSKAYA O, et al.. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs[J]. Chem. Biol., 2004,11(12): 1729-1741. |
| 66 | KIM J N, BLOUNT K F, PUSKARZ I, et al.. Design and antimicrobial action of purine analogues that bind guanine riboswitches[J]. ACS Chem. Biol., 2009,4(11): 915-927. |
| 67 | YAN L H, LE ROUX A, BOYAPELLY K, et al.. Purine analogs targeting the guanine riboswitch as potential antibiotics against Clostridioides difficile [J]. Eur. J. Med. Chem., 2018,143: 755-768. |
| 68 | SERGANOV A, HUANG L, PATEL D J. Structural insights into amino acid binding and gene control by a lysine riboswitch[J]. Nature, 2008,455(7217): 1263-1276. |
| 69 | MUKHERJEE S, BARASH D, SENGUPTA S. Comparative genomics and phylogenomic analyses of lysine riboswitch distributions in bacteria[J/OL]. PLoS ONE, 2017,12(9):e0184314[2021-08-28]. . |
| 70 | ZHOU D, WANG Q, QI Q. Research progress in glmS riboswitch[J]. Acta Microbiol. Sin., 2017,57(8): 1152-1159. |
| 71 | MCCOWN P J, ROTH A, BREAKER R R. An expanded collection and refined consensus model of glmS ribozymes[J]. RNA, 2011,17(4): 728-736. |
| 72 | LUENSE C E, SCHMIDT M S, WITTMANN V, et al.. Carba-sugars activate the glmS-riboswitch of Staphylococcus aureus [J]. ACS Chem. Biol., 2011,6(7): 675-678. |
| 73 | KHAN M A, GOPEL Y, MILEWSKI S, et al.. Two small RNAs conserved in enterobacteriaceae provide intrinsic resistance to antibiotics targeting the cell wall biosynthesis enzyme glucosamine-6-phosphate synthase[J/OL]. Front. Microbiol., 2016,7:908[2021-08-28]. . |
| 74 | FINNEY L A, O'HALLORAN T V. Transition metal speciation in the cell: Insights from the chemistry of metal ion receptors[J]. Sciences, 2003,300(5621): 931-936. |
| 75 | WINKLER W C, BREAKER R R. Regulation of bacterial gene expression by riboswitches[J]. Ann. Rev. Microbiol., 2005,59: 487-517. |
| 76 | DANN C E I, WAKEMAN C A, SIELING C L, et al.. Structure and mechanism of a metal-sensing regulatory RNA[J]. Cell, 2007,130(5): 878-892. |
| 77 | WEINBERG Z, WANG J X, BOGUE J, et al.. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes[J/OL]. Genome Biol., 2010,11(3):R31 [2021-08-28]. . |
| 78 | BAKER J L, SUDARSAN N, WEINBERG Z, et al.. Widespread genetic switches and toxicity resistance proteins for fluoride[J]. Sciences, 2012,335(6065): 233-235. |
| 79 | THAVARAJAH W, SILVERMAN A D, VEROSLOFF M S, et al.. Point-of-use detection of environmental fluoride via a cell-free riboswitch-based biosensor[J]. ACS Synth. Biol., 2020,9(1): 10-18. |
| [1] | Yue SHI, Yao HAN, Hao LI, Yansong SUN. Application Progress of Biosensors Based on Field-effect Transistors in Nucleic Acid Detection [J]. Current Biotechnology, 2025, 15(4): 597-605. |
| [2] | Yuqi YANG, Xiuxia HE. Application of Rolling Circle Amplification Technique in Electrochemical Biosensors [J]. Current Biotechnology, 2023, 13(6): 863-867. |
| [3] | Haitao CAO, Jing ZHU, Haibo ZENG, Yanchen LIU. Research on Feature Selection of Gut Microbiota and Disease Prediction Model Based on Weighted Average [J]. Current Biotechnology, 2023, 13(5): 798-806. |
| [4] | Yunpeng MA, Jing ZHU, Xinghua CUI. Content Estimating of Microbial Dissolved Organic Carbon Based on Machine Learning [J]. Current Biotechnology, 2023, 13(4): 645-653. |
| [5] | Doudou LEI, Runran MA, Jiabo WANG, Weijun KONG. Research Progress on New Biological Detection Technology for Pesticide Residues in Foods [J]. Current Biotechnology, 2023, 13(1): 1-10. |
| [6] | Wenzhuo ZHAO, Chengxun LI, Zuojian HU, Hongxiu YU. Research Progress of Functional Nucleic Acid Used in Pathogenic Bacteria Detection [J]. Current Biotechnology, 2023, 13(1): 30-38. |
| [7] | Yan HU, Ling CHEN. Detection the Affinity Between PDL1 Antibodies and PDL1 Antigen Based on Biolayer Interferometry [J]. Current Biotechnology, 2021, 11(6): 795-801. |
| [8] | BAI Xiaolian, AI Jun*. Application Progress of Biosensor in Detection of Food-borne Pathogen Escherichia coli O157∶H7 [J]. Curr. Biotech., 2021, 11(3): 269-278. |
| [9] | CHEN Shuo1, GAO Jiaqi1, WANG Di2, LONG Yan2, LI Liang2*, ZHANG Xiao1*. DNA Tetrahedral Nanostructure and its Application Progress in Biotechnology [J]. Curr. Biotech., 2020, 10(6): 661-667. |
| [10] | YANG Yuan1§, GUO Yongfu1§, TANG Haozhi1, ZHANG Biao1, SHI Gonglin1, WANG Yating2, YANG Jun2, WEI Lanfang3*. Identification of Pathogen Responsible for Anthracnose Disease of Postharvest Mango in Yunnan Province and its Biocontrol Bacterium Laboratory Screening [J]. Curr. Biotech., 2020, 10(4): 371-377. |
| [11] | ZHANG Hongbing, LIU Hui, SHI Xiuying, LI Huixuan, FAN Daochun. Microalgae Breeding with High Efficient of Oil-producing and Optimization of Cultivation Conditions [J]. Curr. Biotech., 2020, 10(3): 311-319. |
| [12] | DU Zaihui1, LUO Yunbo1, ZHU Longjiao1, XU Wentao1,2*. Special Secondary Structures of DNA and Their Application Progress [J]. Curr. Biotech., 2019, 9(6): 563-570. |
| [13] | LI Kai1,2, LUO Yunbo1,2, XU Wentao1,2*. Research Progress on CRISPR-Cas Mediated Biosensors [J]. Curr. Biotech., 2019, 9(6): 579-591. |
| [14] | ZHANG Yukun1,2, AN Na2, LIU Weixiao2, WAN Yusong2, JIN Wujun2, LI Liang2*, ZHANG Xiao1*. Research Progress of DNA Sensors Based on Surface Plasmon Resonance and Electrochemical Combination [J]. Curr. Biotech., 2019, 9(6): 592-598. |
| [15] | LIN Shenghao1, DU Zaihui1, ZHANG Xiujie2, HUANG Kunlun1,3, LIU Qingliang4, XU Wentao1,3*. Progress on Biosensors Based on Loop-mediated Isothermal Amplification [J]. Curr. Biotech., 2019, 9(6): 599-610. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||