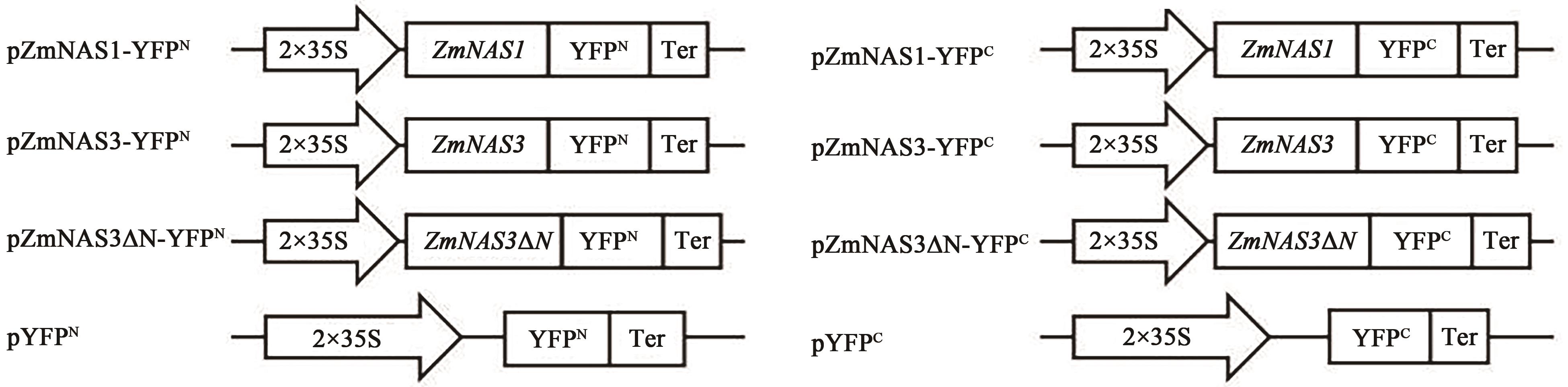
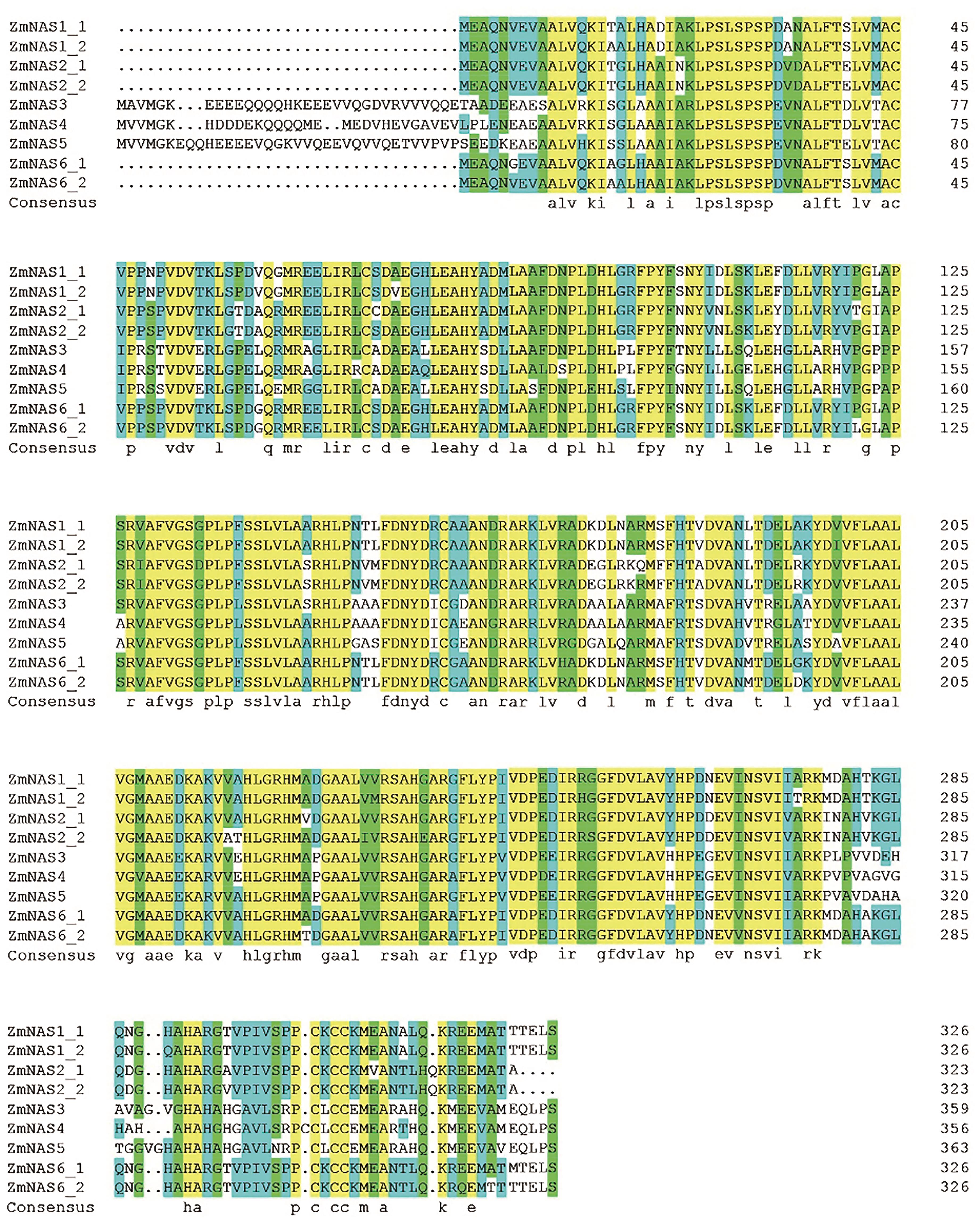
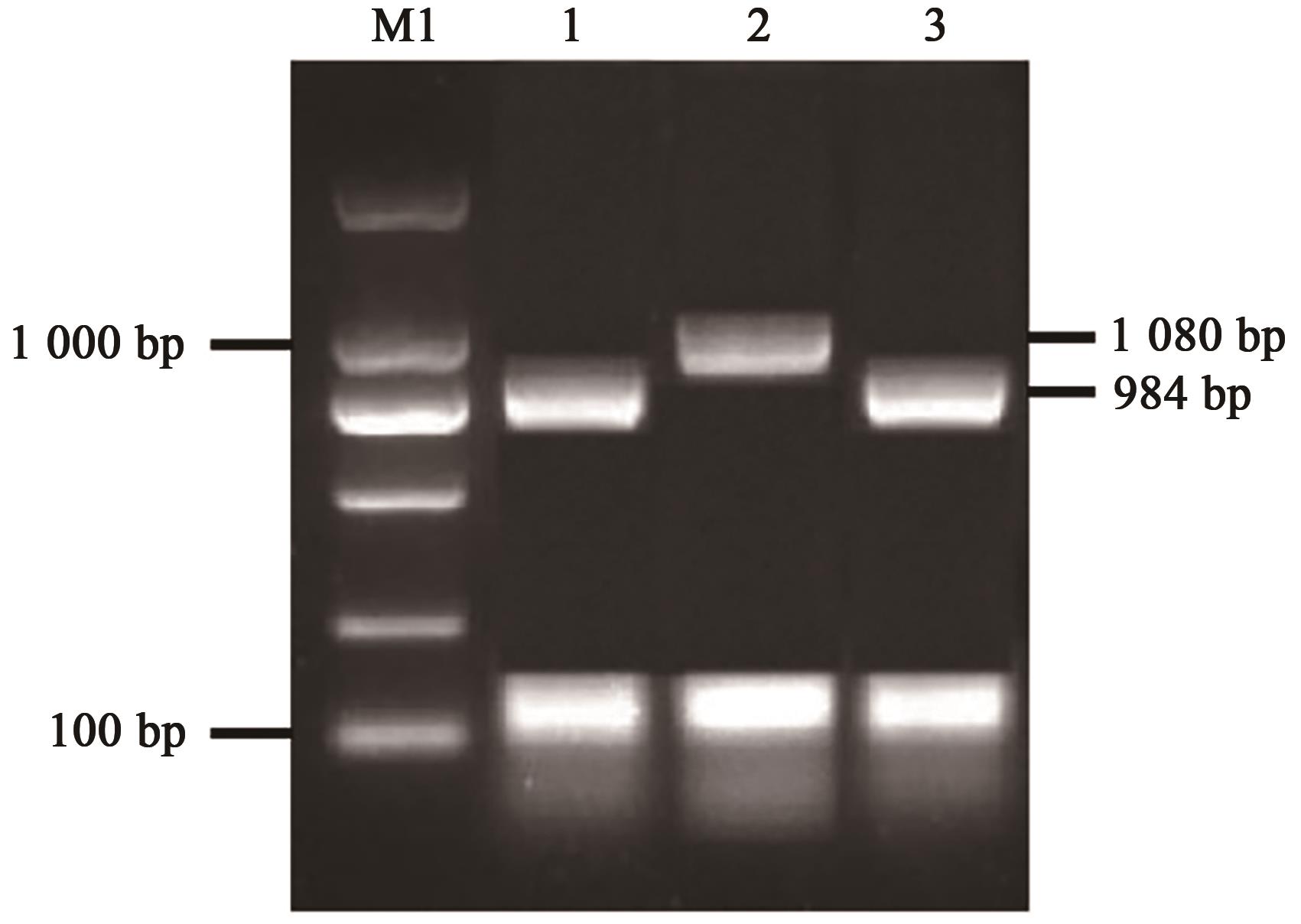
Current Biotechnology ›› 2022, Vol. 12 ›› Issue (5): 728-736.DOI: 10.19586/j.2095-2341.2022.0047
• Articles • Previous Articles Next Articles
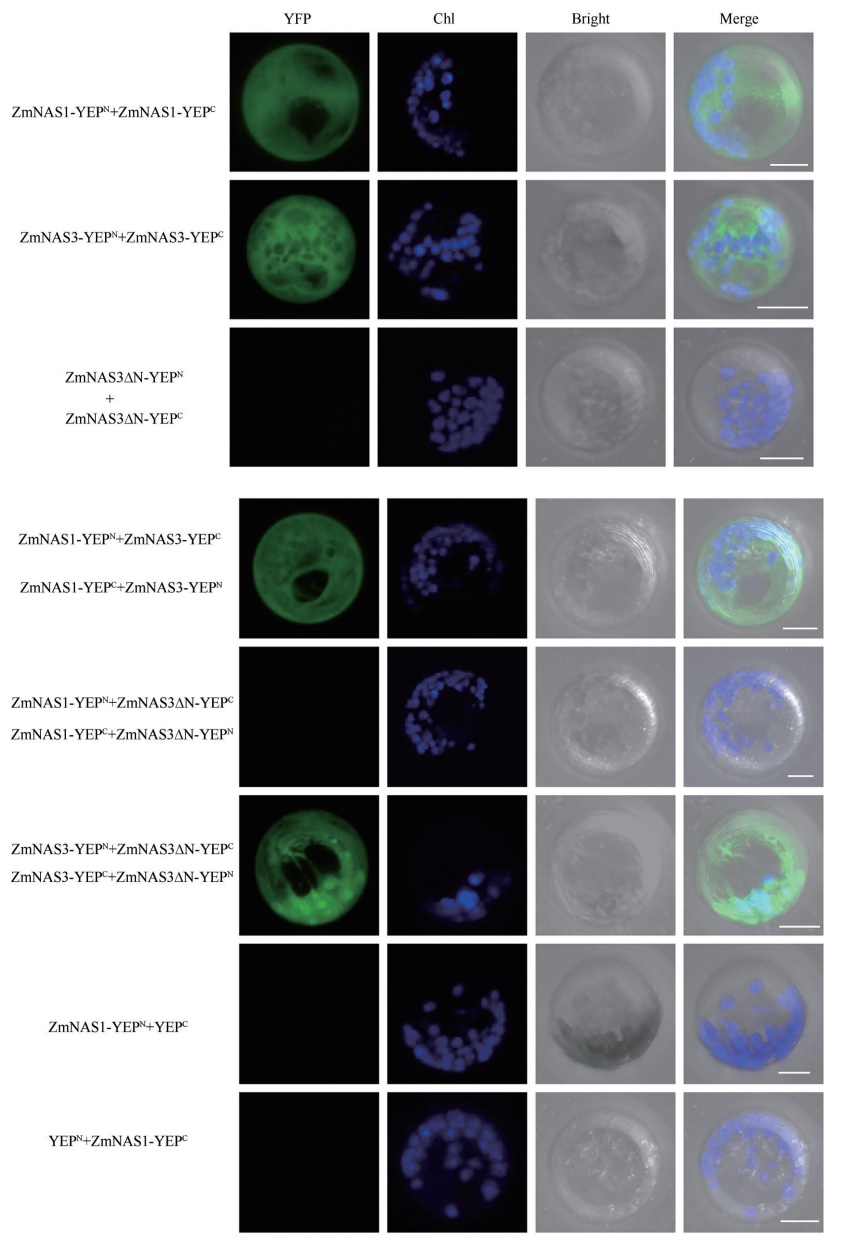
Interactions Between ClassⅠand ClassⅡ NAS Proteins in Maize Using Bimolecular Fluorescence Complementation (BiFC) Assay
Ke XIAO1( ), Xiaojin ZHOU2, Rumei CHEN2, Sen PANG1(
), Xiaojin ZHOU2, Rumei CHEN2, Sen PANG1( )
)
- 1.Department of Applied Chemistry,College of Science,China Agricultural University,Beijing 100193,China
2.Biotechnology Research Institute,Chinese Academy of Agricultural Sciences,Beijing 100081,China
-
Received:2022-03-29Accepted:2022-05-05Online:2022-09-25Published:2022-09-30 -
Contact:Sen PANG
玉米中ClassⅠ和ClassⅡ NAS蛋白之间的BiFC互作研究
- 1.中国农业大学理学院应用化学系,北京 100193
2.中国农业科学院生物技术研究所,北京 100081
-
通讯作者:逄森 -
作者简介:肖克 E-mail:S20193101820@cau.edu.cn; -
基金资助:国家转基因生物新品种培育科技重大专项(2016ZX08003-002)
CLC Number:
Cite this article
Ke XIAO, Xiaojin ZHOU, Rumei CHEN, Sen PANG. Interactions Between ClassⅠand ClassⅡ NAS Proteins in Maize Using Bimolecular Fluorescence Complementation (BiFC) Assay[J]. Current Biotechnology, 2022, 12(5): 728-736.
肖克, 周晓今, 陈茹梅, 逄森. 玉米中ClassⅠ和ClassⅡ NAS蛋白之间的BiFC互作研究[J]. 生物技术进展, 2022, 12(5): 728-736.
share this article
| 基因名称 | 引物名称 | 引物序列(5'→3') |
|---|---|---|
| ZmNAS1 | ZmNAS1-F | 5'-ATGGAGGCCCAGAACGTGGA-3' |
| ZmNAS1-R | 5'-CCACGACAGAGCTGTCCATC-3' | |
| ZmNAS1-NF | 5'-CATTTGGAGAGGACCTCGAGATGGAGGCCCAGAACGTGGA-3' | |
| ZmNAS1-NR | 5'-AACTTTTGCTCCATTCTAGAGATGGACAGCTCTGTCGTGG-3' | |
| ZmNAS1-CF | 5'-CATTTGGAGAGGACCTCGAGATGGAGGCCCAGAACGTGGA-3' | |
| ZmNAS1-CR | 5'-TCGTATGGGTACATTCTAGAGATGGACAGCTCTGTCGTGG-3' | |
| ZmNAS3 | ZmNAS3-F | 5'-CAGCAAAGTGATCAGTGTGA-3' |
| ZmNAS3-R | 5'-CTAGTAGCCAGGTCAATCTG-3' | |
| ZmNAS3-NF | 5'-CATTTGGAGAGGACCTCGAGATGGCCGTCATGGGCAAGGA-3' | |
| ZmNAS3-NR | 5'-AACTTTTGCTCCATTCTAGAGGACGGCAGCTGCTCCATGG-3' | |
| ZmNAS3-CF | 5'-CATTTGGAGAGGACCTCGAGATGGCCGTCATGGGCAAGGA-3' | |
| ZmNAS3-CR | 5'-TCGTATGGGTACATTCTAGAGGACGGCAGCTGCTCCATGG-3' | |
| ZmNAS3∆N-F | 5'-TCTTCACCGACCTCGTCACG-3' | |
| ZmNAS3∆N-R | 5'-CCATGGAGCAGCTGCCGTCC-3' | |
| ZmNAS3∆N-NF | 5'-CATTTGGAGAGGACCTCGAGATGCGCGCGGGCCTCATCCG-3' | |
| ZmNAS3∆N-NR | 5'-AACTTTTGCTCCATTCTAGAGGACGGCAGCTGCTCCATGG-3' | |
| ZmNAS3∆N-CF | 5'-CATTTGGAGAGGACCTCGAGATGCGCGCGGGCCTCATCCG-3' | |
| ZmNAS3∆N-CR | 5'-TCGTATGGGTACATTCTAGAGGACGGCAGCTGCTCCATGG-3' |
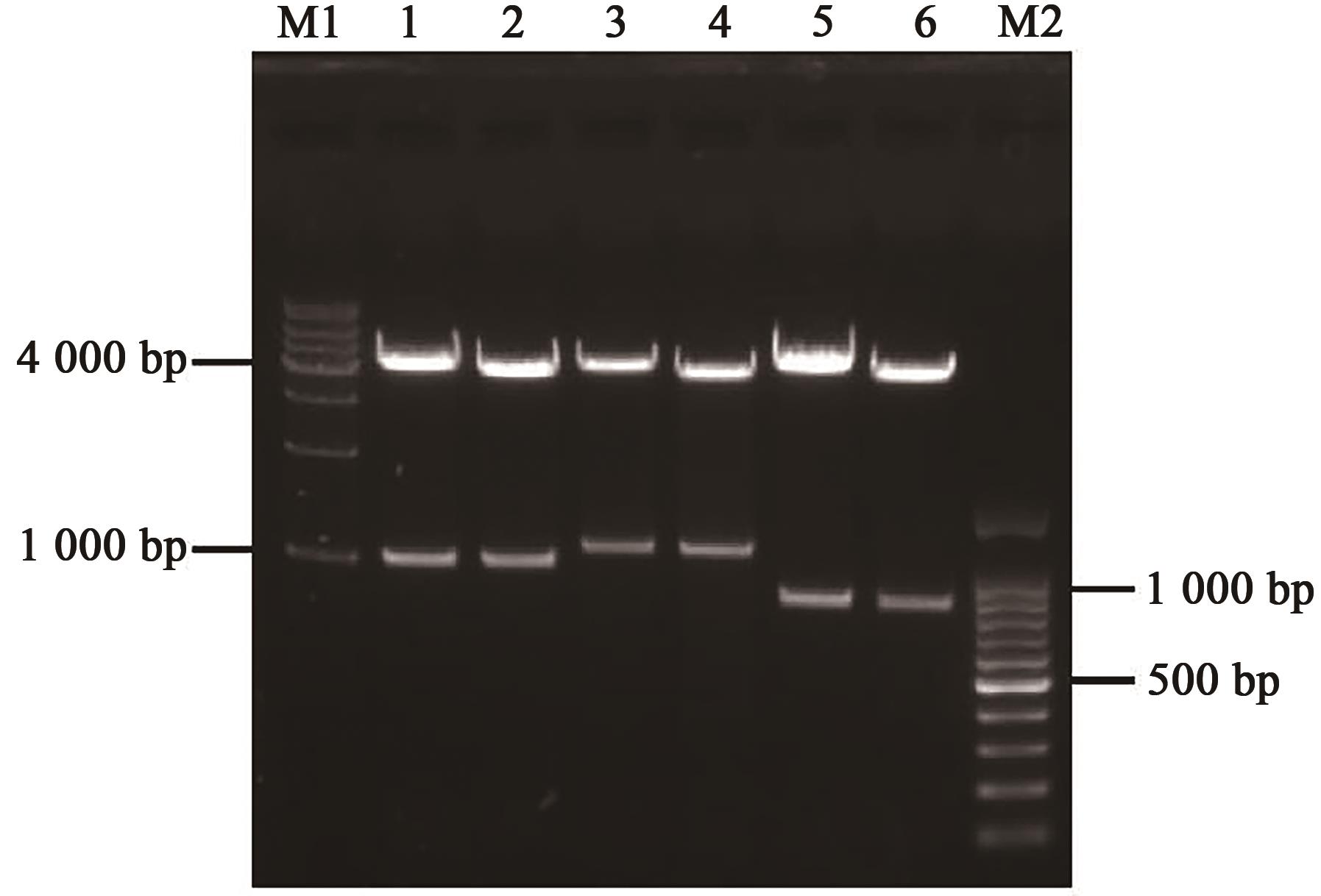
Table 1 Primers used in PCR
| 基因名称 | 引物名称 | 引物序列(5'→3') |
|---|---|---|
| ZmNAS1 | ZmNAS1-F | 5'-ATGGAGGCCCAGAACGTGGA-3' |
| ZmNAS1-R | 5'-CCACGACAGAGCTGTCCATC-3' | |
| ZmNAS1-NF | 5'-CATTTGGAGAGGACCTCGAGATGGAGGCCCAGAACGTGGA-3' | |
| ZmNAS1-NR | 5'-AACTTTTGCTCCATTCTAGAGATGGACAGCTCTGTCGTGG-3' | |
| ZmNAS1-CF | 5'-CATTTGGAGAGGACCTCGAGATGGAGGCCCAGAACGTGGA-3' | |
| ZmNAS1-CR | 5'-TCGTATGGGTACATTCTAGAGATGGACAGCTCTGTCGTGG-3' | |
| ZmNAS3 | ZmNAS3-F | 5'-CAGCAAAGTGATCAGTGTGA-3' |
| ZmNAS3-R | 5'-CTAGTAGCCAGGTCAATCTG-3' | |
| ZmNAS3-NF | 5'-CATTTGGAGAGGACCTCGAGATGGCCGTCATGGGCAAGGA-3' | |
| ZmNAS3-NR | 5'-AACTTTTGCTCCATTCTAGAGGACGGCAGCTGCTCCATGG-3' | |
| ZmNAS3-CF | 5'-CATTTGGAGAGGACCTCGAGATGGCCGTCATGGGCAAGGA-3' | |
| ZmNAS3-CR | 5'-TCGTATGGGTACATTCTAGAGGACGGCAGCTGCTCCATGG-3' | |
| ZmNAS3∆N-F | 5'-TCTTCACCGACCTCGTCACG-3' | |
| ZmNAS3∆N-R | 5'-CCATGGAGCAGCTGCCGTCC-3' | |
| ZmNAS3∆N-NF | 5'-CATTTGGAGAGGACCTCGAGATGCGCGCGGGCCTCATCCG-3' | |
| ZmNAS3∆N-NR | 5'-AACTTTTGCTCCATTCTAGAGGACGGCAGCTGCTCCATGG-3' | |
| ZmNAS3∆N-CF | 5'-CATTTGGAGAGGACCTCGAGATGCGCGCGGGCCTCATCCG-3' | |
| ZmNAS3∆N-CR | 5'-TCGTATGGGTACATTCTAGAGGACGGCAGCTGCTCCATGG-3' |
| 1 | KOBAYASHI T, NISHIZAWA N K. Iron uptake, translocation, and regulation in higher plants[J]. Annu. Rev. Plant Biol., 2012, 63: 131-152. |
| 2 | GUERINOT M L, YI Y. Iron: Nutritious, noxious, and not readily available[J]. Plant Physiol., 1994, 104(3): 815-820. |
| 3 | KOBAYASHIA T, NOZOYEB T, NISHIZAWAA N K. Iron transport and its regulation in plants[J]. Free Radic. Biol. Med., 2019, 133: 11-20. |
| 4 | SENOURA T, SAKASHITA E, KOBAYASHI T, et al.. The iron-chelate transporter OsYSL9 plays a role in iron distribution in developing rice grains[J]. Plant Mol. Biol., 2017, 95(4-5): 375-387. |
| 5 | NOZOYE T, NAGASAKA S, KOBAYASHI T, et al.. Phytosiderophore efflux transporters are crucial for iron acquisition in graminaceous plants[J]. J. Biol. Chem., 2011, 286(7): 5446-5454. |
| 6 | CURIE C, PANAVIENE Z, LOULERGUE C, et al.. Maize yellow stripe1 encodes a membrane protein directly involved in Fe(Ⅲ) uptake[J]. Nature, 2001, 409(6818): 346-349. |
| 7 | CLEMENS S. Metal ligands in micronutrient acquisition and homeostasis[J]. Plant Cell Environ., 2019, 42(10): 2902-2912. |
| 8 | KLATTE M, SCHULER M, WIRTZ M, et al.. The analysis of Arabidopsis nicotianamine synthase mutants reveals functions for nicotianamine in seed iron loading and iron deficiency responses[J]. Plant Physiol., 2009, 150(1): 257-271. |
| 9 | SCHULER M, RELLÁN-ÁLVAREZ R, FINK-STRAUBE C, et al.. Nicotianamine functions in the Phloem-based transport of iron to sink organs, in pollen development and pollen tube growth in Arabidopsis [J]. Plant Cell, 2012, 24(6): 2380-2400. |
| 10 | BECKE R, FRITZ E, MANTEUFFEL R. Subcellular localization and characterization of excessive iron in the nicotianamine-less tomato mutant chloronerva[J]. Plant Physiol., 1995, 108(1): 269-275. |
| 11 | HELL R, STEPHAN U W. Iron uptake, trafficking and homeostasis in plants[J]. Planta, 2003, 216(4): 541-551. |
| 12 | BASHIR K, INOUE H, NAGASAKA S, et al.. Cloning and characterization of deoxymugineic acid synthase genes from graminaceous plants[J]. J. Biol. Chem., 2006, 281(43): 32395-32402. |
| 13 | ZHENG L, CHENG Z, AI C, et al.. Nicotianamine, a novel enhancer of rice iron bioavailability to humans[J/OL]. PLoS ONE, 2010, 5(4): e10190[2022-07-03]. . |
| 14 | EAGLING T, WAWER A A, SHEWRY P R, et al. Iron bioavailability in two commercial cultivars of wheat: comparison between wholegrain and white flour and the effects of nicotianamine and 2'-deoxymugineic acid on iron uptake into Caco-2 cells[J]. J. Agric. Food Chem., 2014, 62(42): 10320-10325. |
| 15 | YOSHIKO M, MASAMI Y, NAHO S, et al.. Iron uptake mediated by the plant-derived chelator nicotianamine in the small intestine[J/OL]. J. Biol. Chem., 2021, 296: 100195[2022-07-03]. . |
| 16 | NOMA M, NOGUCHI M, TAMAKI E. A new amino acid, nicotianamine, from tobacco leaves[J]. Tetrahedron Lett., 1971, 12(22): 2017-2020. |
| 17 | HIGUCHI K, SUZUKI K, NAKANISHI H, et al.. Cloning of nicotianamine synthase genes, novel genes involved in the biosynthesis of phytosiderophores[J]. Plant Physiol., 1999, 119(2): 471-480. |
| 18 | BONNEAU J, BAUMANN U, BEASLEY J, et al.. Identification and molecular characterization of the nicotianamine synthase gene family in bread wheat[J]. Plant Biotechnol. J., 2016, 14(12): 2228-2239. |
| 19 | INOUE H, HIGUCHI K, TAKAHASHI M, et al.. Three rice nicotianamine synthase genes, OsNAS1, OsNAS2, and OsNAS3 are expressed in cells involved in long-distance transport of iron and differentially regulated by iron[J]. Plant J., 2003, 36(3): 366-381. |
| 20 | PEROVIC D, TIFFIN P, DOUCHKOV D, et al.. An integrated approach for the comparative analysis of a multigene family: the nicotianamine synthase genes of barley[J]. Funct. Integr.Genom., 2007, 7(2): 169-179. |
| 21 | DU X, WANG H, HE J, et al.. Identification of nicotianamine synthase genes in Triticum monococcum and their expression under different Fe and Zn concentrations[J]. Gene, 2018, 672: 1-7. |
| 22 | LUDWIG Y, SLAMET-LOEDIN I H. Genetic biofortification to enrich rice and wheat grain iron: from genes to product[J/OL]. Front. Plant Sci., 2019, 10: 833[2022-07-03]. . |
| 23 | ZHOU X, LI S, ZHAO Q, et al. Genome-wide identification, ClassⅠfication and expression profiling of nicotianamine synthase (NAS) gene family in maize[J/OL]. BMC Genom., 2013, 14: 238[2022-07-03]. . |
| 24 | NISHIYAMA R, KATO M, NAGATA S, et al.. Identification of Zn-nicotianamine and Fe-2'-Deoxymugineic acid in the phloem sap from rice plants (Oryza sativa L.)[J]. Plant Cell Physiol., 2012, 53(2): 381-390. |
| 25 | JOHNSON A A, KYRIACOU B, CALLAHAN D L, et al. Constitutive overexpression of the OsNAS gene family reveals single-gene strategies for effective iron- and zinc-biofortification of rice endosperm[J/OL]. PLoS ONE, 2011, 6(9): e24476[2022-07-03]. . |
| 26 | 张鑫,周晓今,逄森,等.玉米ZmENA1基因的克隆及其对金属离子的应答分析[J].生物技术通报,2020,36(11):1-8. |
| 27 | 吕迪,陈茹梅,周晓今.ZmJAZ与ZmMYC2的BiFC互作研究[J].生物技术通报,2022,38(1):77-85. |
| 28 | LING H Q, KOCH G, BÄUMLEIN H, et al.. Map-based cloning of chloronerva, a gene involved in iron uptake of higher plants encoding nicotianamine synthase[J]. Proc. Natl. Acad. Sci. USA, 1999, 96(12): 7098-7103. |
| 29 | SCHÜTZE K, HARTER K, CHABAN C. Post-translational regulation of plant bZIP factors[J]. Trends Plant Sci., 2008, 13(5): 247-255. |
| 30 | LIU B, YANG Z, GOMEZ A, et al.. Signaling mechanisms of plant cryptochromes in Arabidopsis thaliana [J]. J. Plant Res., 2016, 129(2): 137-148. |
| 31 | WANG P, WANG Y, WANG W, et al.. Ubiquitination of phytoene synthase 1 precursor modulates carotenoid biosynthesis in tomato[J/OL]. Commun. Biol., 2020, 3(1): 730[2022-07-03]. . |
| 32 | 孙瑞雪,米薇,叶子弘.基于质谱技术的蛋白质互作研究进展[J].生物技术进展,2022,12(2):161-167. |
| 33 | 王婷,葛怀娜,郭宏.酵母双杂交技术应用进展[J].生物技术进展,2015,5(5):392-396. |
| [1] | Chuancai LIANG, Bo QIU. Echinoside Inhibits IL-1β-induced Chondrocytes Iron Death Through Nrf2/HO-1 Pathway [J]. Current Biotechnology, 2025, 15(4): 720-725. |
| [2] | Yu DING, Bo ZHAO, Jin ZHANG, Xudong GAO. The Role of SIRT1 Deacetylation Modification in Regulating HMGB1-mediated Pyroptosis in Chronic Sinusitis with Nasal Polyps [J]. Current Biotechnology, 2025, 15(3): 535-543. |
| [3] | Zhuoying LIU, Xiaojin ZHOU, Yanli HUANG, Sen PANG. Joint Transcriptome Analysis of Maize Under Salt Stress and MeJA Treatment [J]. Current Biotechnology, 2025, 15(2): 263-275. |
| [4] | HanXiao MA. Development of qPCR Standard Curves for Monitoring Population Dynamics in a Dual-bacterial Fermentative Hydrogen Production System [J]. Current Biotechnology, 2025, 15(2): 325-332. |
| [5] | Qingyun ZHANG, Lei MA, Hua XU, Lian JIN, Junjie ZOU, Baobao WANG, Quanjia CHEN, Miaoyun XU. Mechanism of Lignin Content in Root System Affecting Salt Tolerance in Maize [J]. Current Biotechnology, 2025, 15(1): 67-77. |
| [6] | Xin QI, Xinran LI, Yaning GUO, Dan WANG, Kai LI, Qiong WU, Liang LI. Comparison of Endogenous Genes in Maize Based on Digital PCR [J]. Current Biotechnology, 2025, 15(1): 78-85. |
| [7] | Jingsheng TAN, Guantao ZHAO, Li ZHU, Yubin LI. The Sequence Characterization of Somatic Excision Footprints from Two-element Transposable System of rMrh/Mrh in Maize [J]. Current Biotechnology, 2024, 14(4): 601-609. |
| [8] | Yan ZENG, Hengcheng ZHU, Kang YANG. The Mechanism of DNASE1L3 in Renal Cell Carcinoma [J]. Current Biotechnology, 2024, 14(3): 486-491. |
| [9] | Jiaqi SUN, Jia GUO, Chuang ZHANG, Qing LIU, Ziyu WANG, Hanchao XIA, Buxuan QIAN, Fangfang ZHAO, Qi WANG, Jianfeng LIU, Xiangguo LIU. Research Progress of Phosphite Dehydrogenase in Genetically Engineered Microorganisms and Plants [J]. Current Biotechnology, 2024, 14(2): 173-181. |
| [10] | Guantao ZHAO, Jingsheng TAN, Li ZHU, Yubin LI. The Sequence Characterization of the Somatic Footprints upon the Transposition of Non-autonomous Transposon rDt in Maize [J]. Current Biotechnology, 2024, 14(2): 248-256. |
| [11] | Lingyan LI, Bing XIAO, Xudong ZHANG, Hua ZHANG, Ziyan CHEN, Haoqian WANG, Xiujie ZHANG, Hong CHEN, Jingang LIANG. Establishment and Standardization of Real-time PCR Method for Qualitative Detection of Genetically Modified Maize MON87411 [J]. Current Biotechnology, 2024, 14(2): 257-262. |
| [12] | Xiaoxue WANG, Yi JI, Huiru YU, Junfeng XU, Cheng PENG, Xiaofu WANG, Yueying LI, Xiaoyun CHEN. A Rapid Method for Bovine-derived Components Based on Recombinase Polymerase Amplification Technology [J]. Current Biotechnology, 2024, 14(2): 278-286. |
| [13] | Caihong WANG, Yuhan SHAO, Mengyuan JIANG. The Mechanisms of IL-17 Inhibiting Pseudomonas aeruginosa Infection on Lung Epithelial Cells [J]. Current Biotechnology, 2024, 14(2): 304-311. |
| [14] | Xiaohan MA, Lijing HUANG, Fei WANG, Chenhui LI. A New Perspective to Evaluate the Treatment of Rheumatoid Arthritis with Biological Products [J]. Current Biotechnology, 2024, 14(1): 55-59. |
| [15] | Zhenhua KE. Research on Detection Methods of Pseudomonas aeruginosa in Cold Chain Food and Construction of Evolutionary Tree [J]. Current Biotechnology, 2023, 13(6): 940-944. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||