| 1 |
CHEN Z, ZHANG Y. Role of mammalian DNA methyltransferases in development[J]. Annu. Rev. Biochem., 2020, 89: 135-158.
|
| 2 |
KUZNETSOV N A, KANAZHEVSKAYA L Y, FEDOROVA O S. DNA demethylation in the processes of repair and epigenetic regulation performed by 2-Ketoglutarate-dependent DNA dioxygenases[J/OL]. Int. J. Mol. Sci., 2021, 22(19): 10540 [2022-01-07]. .
|
| 3 |
SMALLWOOD S A, KELSEY G. De novo DNA methylation: A germ cell perspective[J]. Trends Genet., 2012, 28(1): 33-42.
|
| 4 |
HAJKOVA P, ANCELIN K, WALDMANN T, et al.. Chromatin dynamics during epigenetic reprogramming in the mouse germ line[J]. Nature, 2008, 452(7189): 877-881.
|
| 5 |
HAJKOVA P. Epigenetic reprogramming--taking a lesson from the embryo[J]. Curr. Opin. Cell Biol., 2010, 22(3): 342-350.
|
| 6 |
IYER L M, ZHANG D, DE SOUZA R F, et al.. Lineage-specific expansions of TET/JBP genes and a new class of DNA transposons shape fungal genomic and epigenetic landscapes[J]. Proc. Natl. Acad. Sci. USA, 2014, 111(5): 1676-1683.
|
| 7 |
ZHU F, ZHU Q, YE D, et al.. Sin3a-Tet1 interaction activates gene transcription and is required for embryonic stem cell pluripotency[J]. Nucl. Acids Res., 2018, 46(12): 6026-6040.
|
| 8 |
LI X, YUE X, PASTOR W A, et al.. Tet proteins influence the balance between neuroectodermal and mesodermal fate choice by inhibiting Wnt signaling[J/OL]. Proc. Natl. Acad. Sci. USA, 2016, 113(51): E8267-E8276 [2022-06-11]. .
|
| 9 |
KOIVUNEN P, LAUKKA T. The TET enzymes[J]. Cell Mol. Life Sci., 2018, 75(8): 1339-1348.
|
| 10 |
MAHAJAN V, FARQUHAR C, PONNAMPALAM A P. Could DNA hydroxymethylation be crucial in influencing steroid hormone signaling in endometrial biology and endometriosis?[J]. Mol. Reprod. Dev., 2020, 87(1): 7-16.
|
| 11 |
SARKAR D, LEUNG E Y, BAGULEY B C, et al.. Epigenetic regulation in human melanoma: past and future[J]. Epigenetics, 2015, 10(2): 103-121.
|
| 12 |
TERRADAS-TERRADAS M, ROBERTSON N A, CHANDRA T, et al.. Clonality in haematopoietic stem cell ageing[J/OL]. Mech. Ageing Dev., 2020, 189: 111279 [2021-12-05]. .
|
| 13 |
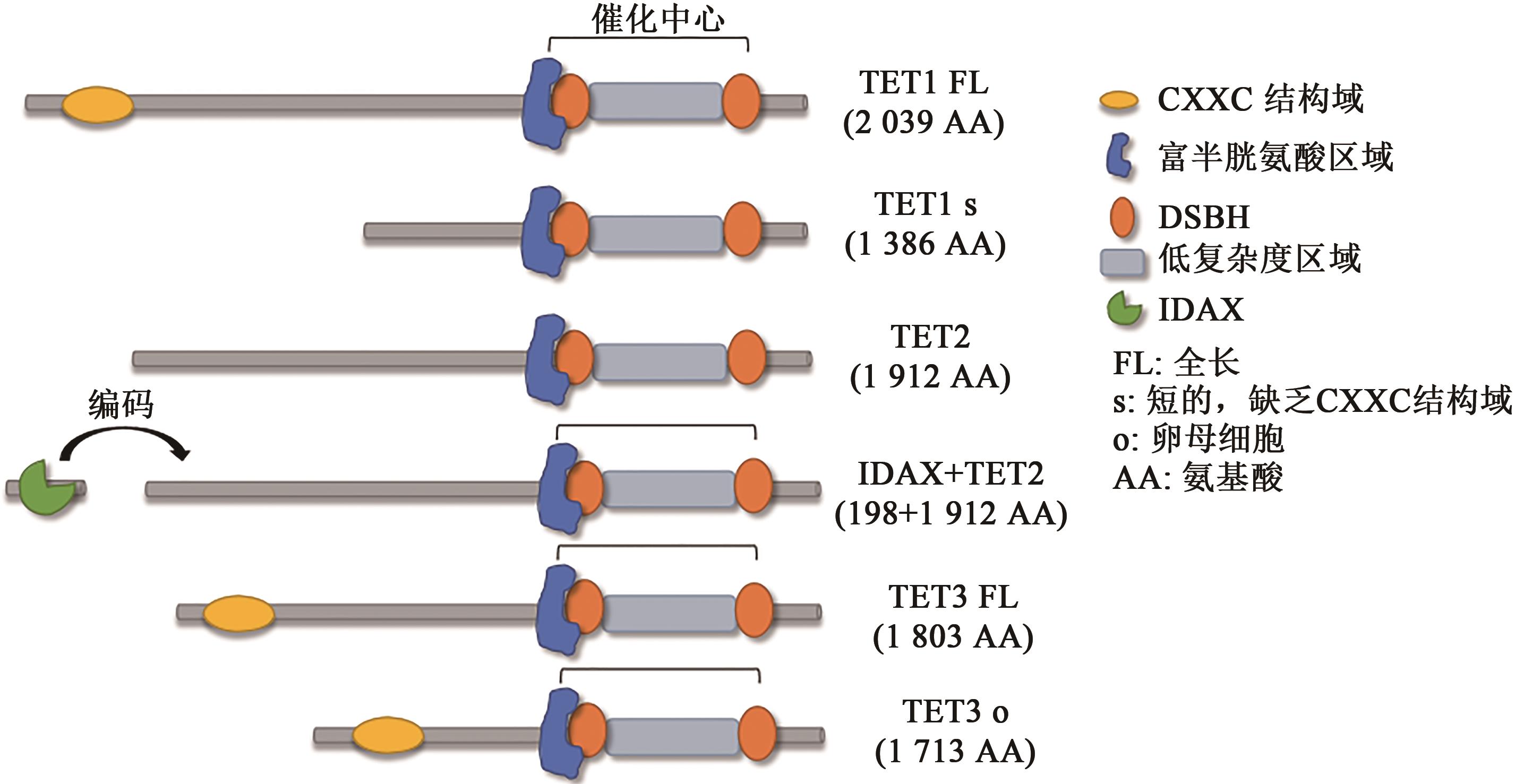
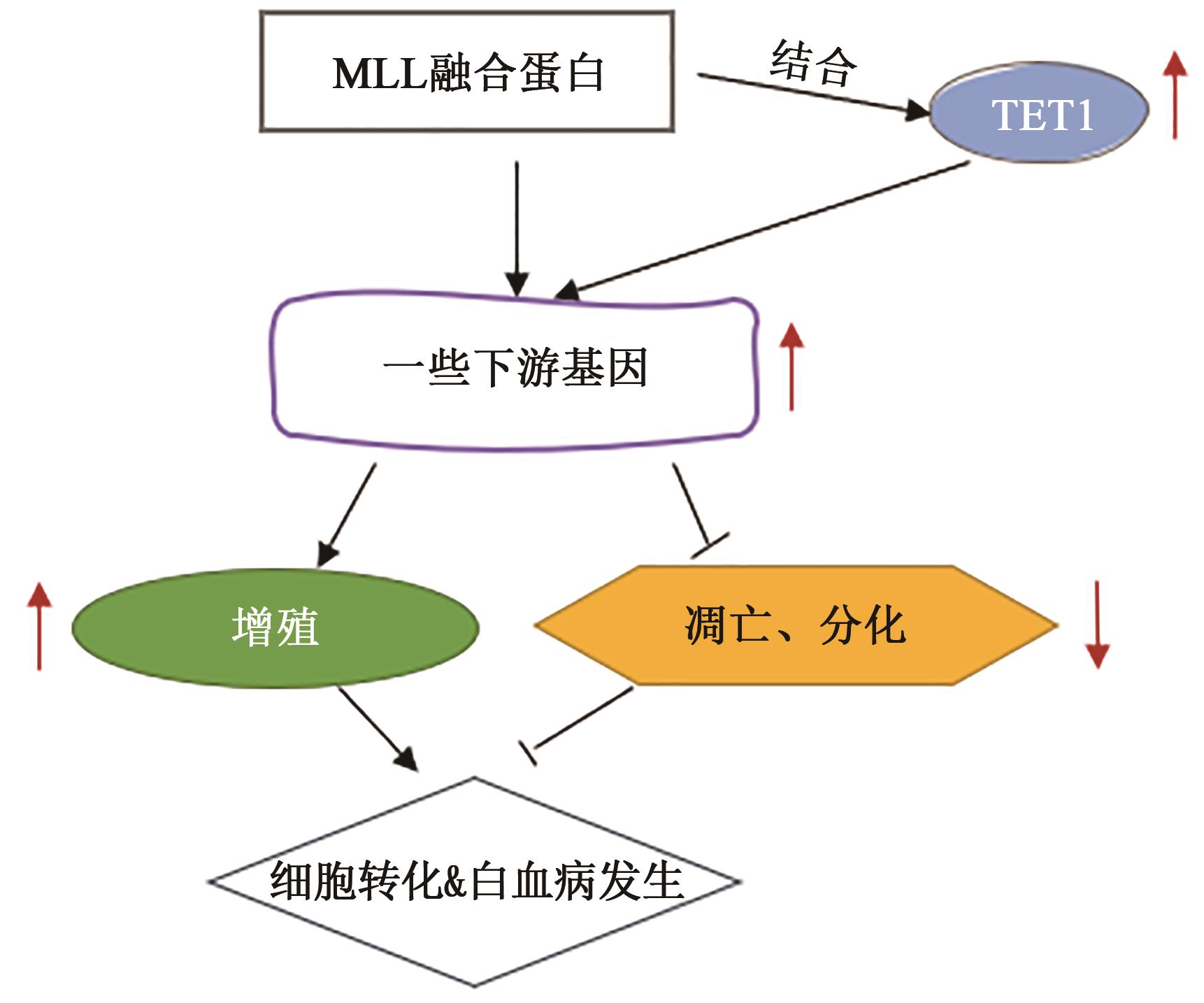
LORSBACH R B, MOORE J, MATHEW S, et al.. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23)[J]. Leukemia, 2003, 17(3): 637-641.
|
| 14 |
ROSS S E, BOGDANOVIC O. TET enzymes, DNA demethylation and pluripotency[J]. Biochem. Soc. Trans., 2019, 47(3): 875-885.
|
| 15 |
HU L, LI Z, CHENG J, et al.. Crystal structure of TET2-DNA complex: insight into TET-mediated 5mC oxidation[J]. Cell, 2013, 155(7): 1545-1555.
|
| 16 |
MELAMED P, YOSEFZON Y, DAVID C, et al.. Tet enzymes, variants, and differential effects on function[J/OL]. Front. Cell Dev. Biol., 2018, 6: 22 [2021-12-05]. .
|
| 17 |
TAHILIANI M, KOH K P, SHEN Y, et al.. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1[J]. Science, 2009, 324(5929): 930-935.
|
| 18 |
WU S C, ZHANG Y. Active DNA demethylation: many roads lead to Rome[J]. Nat. Rev. Mol. Cell Biol., 2010, 11(9): 607-620.
|
| 19 |
BOCHTLER M, KOLANO A, XU G L. DNA demethylation pathways: additional players and regulators[J]. Bioessays, 2017, 39(1): 1-13.
|
| 20 |
LIU Z J, MARTINEZ C S, DELFT P, et al.. Sequencing abasic sites in DNA at single-nucleotide resolution[J]. Nat. Chem., 2019, 11(7): 629-637.
|
| 21 |
GUO F, LI X, LIANG D, et al.. Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote[J]. Cell Stem Cell, 2014, 15(4): 447-459.
|
| 22 |
HU L, LU J, CHENG J, et al.. Structural insight into substrate preference for TET-mediated oxidation[J]. Nature, 2015, 527(7576): 118-122.
|
| 23 |
LI W, LIU M. Distribution of 5-hydroxymethylcytosine in different human tissues[J/OL]. J. Nucl. Acids, 2011, 2011: 870726 [2021-12-06]. .
|
| 24 |
GLOBISCH D, MUNZEL M, MULLER M, et al.. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates[J/OL]. PLoS ONE, 2010, 5(12): e15367 [2021-12-06]. .
|
| 25 |
JIN S G, WU X, LI A X, et al.. Genomic mapping of 5-hydroxymethylcytosine in the human brain[J]. Nucl. Acids Res., 2011, 39(12): 5015-5024.
|
| 26 |
CAI J, CHEN L, ZHANG Z, et al.. Genome-wide mapping of 5-hydroxymethylcytosines in circulating cell-free DNA as a non-invasive approach for early detection of hepatocellular carcinoma[J]. Gut, 2019, 68(12): 2195-2205.
|
| 27 |
BENESOVA M, TREJBALOVA K, KUCEROVA D, et al.. Overexpression of TET dioxygenases in seminomas associates with low levels of DNA methylation and hydroxymethylation[J]. Mol. Carcinog., 2017, 56(8): 1837-1850.
|
| 28 |
ONO R, TAKI T, TAKETANI T, et al.. LCX, Leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23)[J]. Cancer Res., 2002, 62(14): 4075-4080.
|
| 29 |
HUANG H, JIANG X, LI Z, et al.. TET1 plays an essential oncogenic role in MLL-rearranged leukemia[J]. Proc. Natl. Acad Sci. USA, 2013, 110(29): 11994-11999.
|
| 30 |
SEETHY A, PETHUSAMY K, CHATTOPADHYAY I, et al.. TETology: epigenetic mastermind in action[J]. Appl. Bioche Biotechnol., 2021, 193(6): 1701-1726.
|
| 31 |
MORAN-CRUSIO K, REAVIE L, SHIH A, et al.. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation[J]. Cancer Cell, 2011, 20(1): 11-24.
|
| 32 |
TEFFERI A, LIM K H, ABDEL-WAHAB O, et al.. Detection of mutant TET2 in myeloid malignancies other than myeloproliferative neoplasms: CMML, MDS, MDS/MPN and AML[J]. Leukemia, 2009, 23(7): 1343-1355.
|
| 33 |
WANG Y, XIAO M, CHEN X, et al.. WT1 recruits TET2 to regulate its target gene expression and suppress leukemia cell proliferation[J]. Mol. Cell, 2015, 57(4): 662-673.
|
| 34 |
PAN F, WINGO T S, ZHAO Z, et al.. Tet2 loss leads to hypermutagenicity in haematopoietic stem/progenitor cells[J/OL]. Nat. Commun., 2017, 8: 15102 [2021-12-05]. .
|
| 35 |
PULIKKOTTIL A J, BAMEZAI S, AMMER T, et al.. TET3 promotes AML growth and epigenetically regulates glucose metabolism and leukemic stem cell associated pathways[J]. Leukemia, 2022, 36(2): 416-425.
|
| 36 |
SZULWACH K E, LI X, LI Y, et al.. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells[J/OL]. PLoS Genet., 2011, 7(6): e1002154 [2021-12-15]. .
|
| 37 |
MACARTHUR I C, DAWLATY M M. TET Enzymes and 5-Hydroxymethylcytosine in Neural Progenitor Cell Biology and Neurodevelopment[J/OL]. Front. Cell Dev. Biol., 2021, 9: 645335 [2022-02-05]. .
|
| 38 |
LIAN H, LI W B, JIN W L. The emerging insights into catalytic or non-catalytic roles of TET proteins in tumors and neural development[J]. Oncotarget, 2016, 7(39): 64512-64525.
|
| 39 |
RUDENKO A, DAWLATY M M, SEO J, et al.. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction[J]. Neuron, 2013, 79(6): 1109-1122.
|
| 40 |
DAWLATY M M, BREILING A, LE T, et al.. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development[J]. Dev. Cell, 2013, 24(3): 310-323.
|
| 41 |
VERMA N, PAN H, DORE L C, et al.. TET proteins safeguard bivalent promoters from de novo methylation in human embryonic stem cells[J]. Nat. Genet., 2018, 50(1): 83-95.
|
| 42 |
PALMER A J, SAVERY D, MASSA V, et al.. Genetic interaction of Pax3 mutation and canonical Wnt signaling modulates neural tube defects and neural crest abnormalities[J/OL]. Genesis, 2021, 59(11): e23445 [2022-02-04]. .
|
| 43 |
LU X L, WANG L, CHANG S Y, et al.. Sonic hedgehog signaling affected by promoter hypermethylation induces aberrant Gli2 expression in spina Bifida[J]. Mol. Neurobiol., 2016, 53(8): 5413-5424.
|
| 44 |
FENG J, PENA C J, PURUSHOTHAMAN I, et al.. Tet1 in nucleus accumbens opposes depression- and anxiety-like behaviors[J]. Neuropsychopharmacology, 2017, 42(8): 1657-1669.
|
| 45 |
COCHRAN J N, GEIER E G, BONHAM L W, et al.. Non-coding and loss-of-function coding variants in TET2 are associated with multiple neurodegenerative diseases[J]. Am. J. Hum. Genet., 2020, 106(5): 632-645.
|
| 46 |
BECK D B, PETRACOVICI A, HE C, et al.. Delineation of a human mendelian disorder of the DNA demethylation machinery: TET3 deficiency[J]. Am. J. Hum. Genet., 2020, 106(2): 234-45.
|
| 47 |
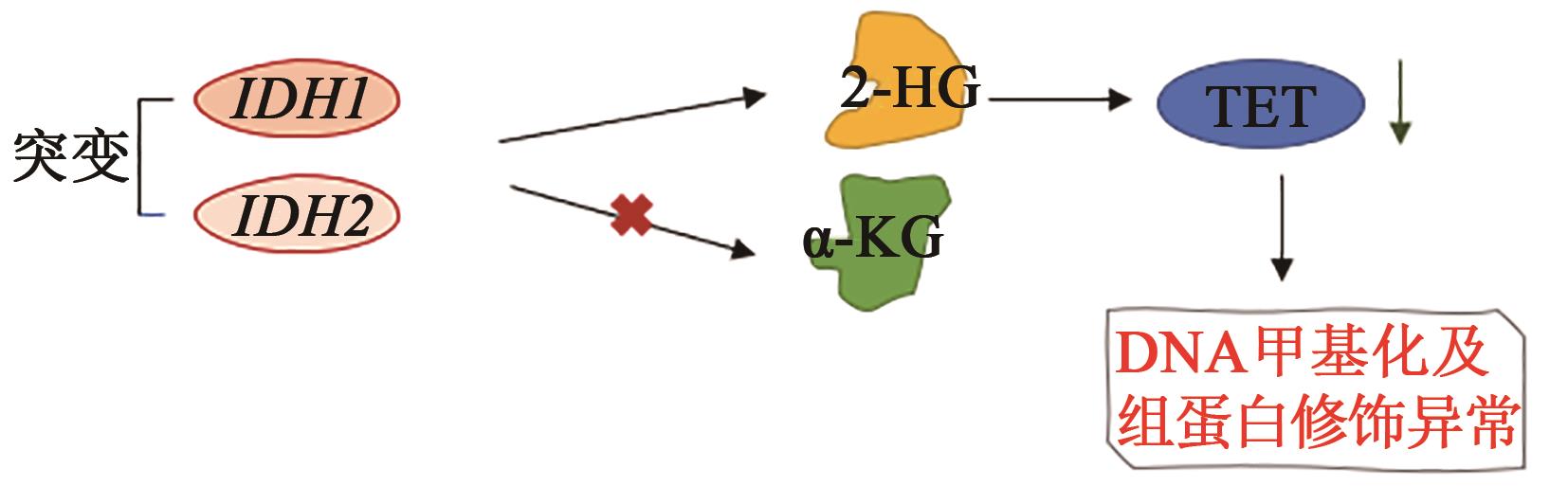
XU W, YANG H, LIU Y, et al.. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases[J]. Cancer Cell, 2011, 19(1): 17-30.
|
| 48 |
CARELLA A, TEJEDOR J R, GARCIA M G, et al.. Epigenetic downregulation of TET3 reduces genome-wide 5hmC levels and promotes glioblastoma tumorigenesis[J]. Int. J. Cancer, 2020, 146(2): 373-387.
|
| 49 |
BRAY J K, DAWLATY M M, VERMA A, et al.. Roles and regulations of TET enzymes in solid tumors[J]. Trends Cancer, 2021, 7(7): 635-646.
|
| 50 |
GARCíA M G, CARELLA A, URDINGUIO R G, et al.. Epigenetic dysregulation of TET2 in human glioblastoma[J]. Oncotarget, 2018, 9(40): 25922-25934.
|
| 51 |
RAWLUSZKO-WIECZOREK A A, SIERA A, JAGODZINSKI P P. TET proteins in cancer: current ‘state of the art’[J]. Crit. Rev. Oncol. Hematol., 2015, 96(3): 425-436.
|
| 52 |
RASMUSSEN K D, HELIN K. Role of TET enzymes in DNA methylation, development, and cancer[J]. Genes Dev., 2016, 30(7): 733-750.
|
| 53 |
SHEN G, SHEN H, ZHANG J, et al.. DNA methylation in Hepatoblastoma-a literature review[J/OL]. Ital. J. Pediatr., 2020, 46(1): 113 [2021-12-09]. .
|
| 54 |
ZHANG J, HAN X, GAO C, et al.. 5-Hydroxymethylome in circulating cell-free DNA as a potential biomarker for non-small-cell lung cancer[J]. Genom. Proteom. Bioinform., 2018, 16(3): 187-199.
|
| 55 |
TSAGARATOU A, LIO C J, YUE X, et al.. TET Methylcytosine Oxidases in T Cell and B cell development and function[J/OL]. Front. Immunol., 2017, 8: 220 [2021-12-15]. .
|
| 56 |
APPLEBAUM M A, BARR E K, KARPUS J, et al.. 5-Hydroxymethylcytosine profiles are prognostic of outcome in neuroblastoma and reveal transcriptional networks that correlate with tumor phenotype[J/OL]. JCO Precis. Oncol., 2019, 3: PO.18.00402 [2021-12-08]. .
|
| 57 |
HE Y F, LI B Z, LI Z, et al.. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA[J]. Science, 2011, 333(6047): 1303-1307.
|
| 58 |
TIE J, COHEN J D, WANG Y, et al.. Circulating Tumor dna analyses as markers of recurrence risk and benefit of adjuvant therapy for stage Ⅲ colon cancer[J]. JAMA Oncol., 2019, 5(12): 1710-1717.
|
| 59 |
XU T, GAO H. Hydroxymethylation and tumors: can 5-hydroxymethylation be used as a marker for tumor diagnosis and treatment?[J/OL]. Hum. Genomics, 2020, 14(1): 15 [2022-01-07]. .
|
 ), Xuejia HE1,2, Fan LIU1,3, Pei PEI1, Shan WANG1(
), Xuejia HE1,2, Fan LIU1,3, Pei PEI1, Shan WANG1( )
)
 ), 何学佳1,2, 刘帆1,3, 裴培1, 王珊1(
), 何学佳1,2, 刘帆1,3, 裴培1, 王珊1( )
)