Current Biotechnology ›› 2025, Vol. 15 ›› Issue (1): 78-85.DOI: 10.19586/j.2095-2341.2023.0165
• Articles • Previous Articles Next Articles
Comparison of Endogenous Genes in Maize Based on Digital PCR
Xin QI1( ), Xinran LI1, Yaning GUO2, Dan WANG1, Kai LI1, Qiong WU3, Liang LI1(
), Xinran LI1, Yaning GUO2, Dan WANG1, Kai LI1, Qiong WU3, Liang LI1( )
)
- 1.Institute of Quality Standard and Testing Technology for Agro-Products of Chinese Academy of Agricultural Sciences,Beijing 100081,China
2.College of Food Science and Bioengineering,Tianjin Agricultural University,Tianjin 300384,China
3.Beijing Genseed Technology Co. ,Ltd. ,Beijing 100194,China
-
Received:2023-12-18Accepted:2024-10-31Online:2025-01-25Published:2025-03-07 -
Contact:Liang LI
基于数字PCR方法的玉米内标准基因比较研究
齐鑫1( ), 李欣然1, 郭亚宁2, 王丹1, 李凯1, 吴琼3, 李亮1(
), 李欣然1, 郭亚宁2, 王丹1, 李凯1, 吴琼3, 李亮1( )
)
- 1.中国农业科学院农业质量标准与检测技术研究所,北京 100081
2.天津农学院食品科学与生物工程学院,天津 300384
3.北京创种科技有限公司,北京 100194
-
通讯作者:李亮 -
作者简介:齐鑫 E-mail: qixin_0908@163.com; -
基金资助:农业生物育种国家科技重大专项(2022ZD04020)
CLC Number:
Cite this article
Xin QI, Xinran LI, Yaning GUO, Dan WANG, Kai LI, Qiong WU, Liang LI. Comparison of Endogenous Genes in Maize Based on Digital PCR[J]. Current Biotechnology, 2025, 15(1): 78-85.
齐鑫, 李欣然, 郭亚宁, 王丹, 李凯, 吴琼, 李亮. 基于数字PCR方法的玉米内标准基因比较研究[J]. 生物技术进展, 2025, 15(1): 78-85.
share this article
| 基因名称 | 引物探针序列(5'→3') | 扩增子/bp | 参考文献 | |
|---|---|---|---|---|
| hmg | F-1 | 5′-TTGGACTAGAAATCTCGTGCTGA-3′ | 79 | [ |
| R-1 | 5′-GCTACATAGGGAGCCTTGTCCT-3′ | |||
| P-1 | 5′-CAATCCACACAAACGCACGCGTA-3′ | |||
| adh1-1 | F-1 | 5′-CGTCGTTTCCCATCTCTTCCTCC-3′ | 135 | [ |
| R-1 | 5′-CCACTCCGAGACCCTCAGTC-3′ | |||
| P-1 | 5′-AATCAGGGCTCATTTTCTCGCTCCTCA-3′ | |||
| adh1-2 | F-2 | 5′-CCTTCTTGGCGGCTTATCTG-3′ | 70 | [ |
| R-2 | 5′-CCAGCCTCATGGCCAAAG-3′ | |||
| P-2 | 5′-CTTAGGGGCAGACTCCCGTGTTCCCT-3′ | |||
| ivr1-1 | F-1 | 5′-CGCTCTGTACAAGCGTGC-3′ | 104 | [ |
| R-1 | 5′-GCAAAGTGTTGTGCTTGGACC-3′ | |||
| P-1 | 5′-CACGTGAGAATTTCCGTCTACTCGAGCCT-3′ | |||
| ivr1-2 | F-2 | 5′-TGGCGGACGACGACTTGT-3′ | 79 | [ |
| R-2 | 5′-AAAGTTTGGAGGCTGCCGT-3′ | |||
| P-2 | 5′-CGAGCAGACCGCCGTGTACTTCTACC-3′ | |||
| zein-1 | F-1 | 5′-CGTGTCCGTCCCTGATGC-3′ | 83 | [ |
| R-1 | 5′-AGGCGTCATCATCTGTGGC-3′ | |||
| P-1 | 5′-CAACTGTTGGCCTTACCGCTTCAGACG-3′ | |||
| zein-2 | F-2 | 5′-GCCATTGGGTACCATGAACC-3′ | 104 | [ |
| R-2 | 5′-AGGCCAACAGTTGCTGCAG-3′ | |||
| P-2 | 5′-AGCTTGATGGCGTGTCCGTCCCT-3′ | |||
| zSSIIb-1 | F-1 | 5′-CTCCCAATCCTTTGACATCTGC-3′ | 151 | [ |
| R-1 | 5′-TCGATTTCTCTCTTGGTGACAGG-3′ | |||
| P-1 | 5′-AGCAAAGTCAGAGCGCTGCAATGCA-3′ | |||
| zSSIIb-2 | F-2 | 5′-CCAATCCTTTGACATCTGCTCC-3′ | 114 | [ |
| R-2 | 5′-GATCAGCTTTGGGTCCGGA-3′ | |||
| P-2 | 5′-AGCAAAGTCAGAGCGCTGCAATGCA-3′ | |||
Table 1 Primer probe sequences of endogenous genes in maize
| 基因名称 | 引物探针序列(5'→3') | 扩增子/bp | 参考文献 | |
|---|---|---|---|---|
| hmg | F-1 | 5′-TTGGACTAGAAATCTCGTGCTGA-3′ | 79 | [ |
| R-1 | 5′-GCTACATAGGGAGCCTTGTCCT-3′ | |||
| P-1 | 5′-CAATCCACACAAACGCACGCGTA-3′ | |||
| adh1-1 | F-1 | 5′-CGTCGTTTCCCATCTCTTCCTCC-3′ | 135 | [ |
| R-1 | 5′-CCACTCCGAGACCCTCAGTC-3′ | |||
| P-1 | 5′-AATCAGGGCTCATTTTCTCGCTCCTCA-3′ | |||
| adh1-2 | F-2 | 5′-CCTTCTTGGCGGCTTATCTG-3′ | 70 | [ |
| R-2 | 5′-CCAGCCTCATGGCCAAAG-3′ | |||
| P-2 | 5′-CTTAGGGGCAGACTCCCGTGTTCCCT-3′ | |||
| ivr1-1 | F-1 | 5′-CGCTCTGTACAAGCGTGC-3′ | 104 | [ |
| R-1 | 5′-GCAAAGTGTTGTGCTTGGACC-3′ | |||
| P-1 | 5′-CACGTGAGAATTTCCGTCTACTCGAGCCT-3′ | |||
| ivr1-2 | F-2 | 5′-TGGCGGACGACGACTTGT-3′ | 79 | [ |
| R-2 | 5′-AAAGTTTGGAGGCTGCCGT-3′ | |||
| P-2 | 5′-CGAGCAGACCGCCGTGTACTTCTACC-3′ | |||
| zein-1 | F-1 | 5′-CGTGTCCGTCCCTGATGC-3′ | 83 | [ |
| R-1 | 5′-AGGCGTCATCATCTGTGGC-3′ | |||
| P-1 | 5′-CAACTGTTGGCCTTACCGCTTCAGACG-3′ | |||
| zein-2 | F-2 | 5′-GCCATTGGGTACCATGAACC-3′ | 104 | [ |
| R-2 | 5′-AGGCCAACAGTTGCTGCAG-3′ | |||
| P-2 | 5′-AGCTTGATGGCGTGTCCGTCCCT-3′ | |||
| zSSIIb-1 | F-1 | 5′-CTCCCAATCCTTTGACATCTGC-3′ | 151 | [ |
| R-1 | 5′-TCGATTTCTCTCTTGGTGACAGG-3′ | |||
| P-1 | 5′-AGCAAAGTCAGAGCGCTGCAATGCA-3′ | |||
| zSSIIb-2 | F-2 | 5′-CCAATCCTTTGACATCTGCTCC-3′ | 114 | [ |
| R-2 | 5′-GATCAGCTTTGGGTCCGGA-3′ | |||
| P-2 | 5′-AGCAAAGTCAGAGCGCTGCAATGCA-3′ | |||
| 基因名称 | 59.0 ℃ | 58.6 ℃ | 58.0 ℃ | 57.1 ℃ | 55.9 ℃ | 55.0 ℃ | 平均值±标准差 | RSD/% |
|---|---|---|---|---|---|---|---|---|
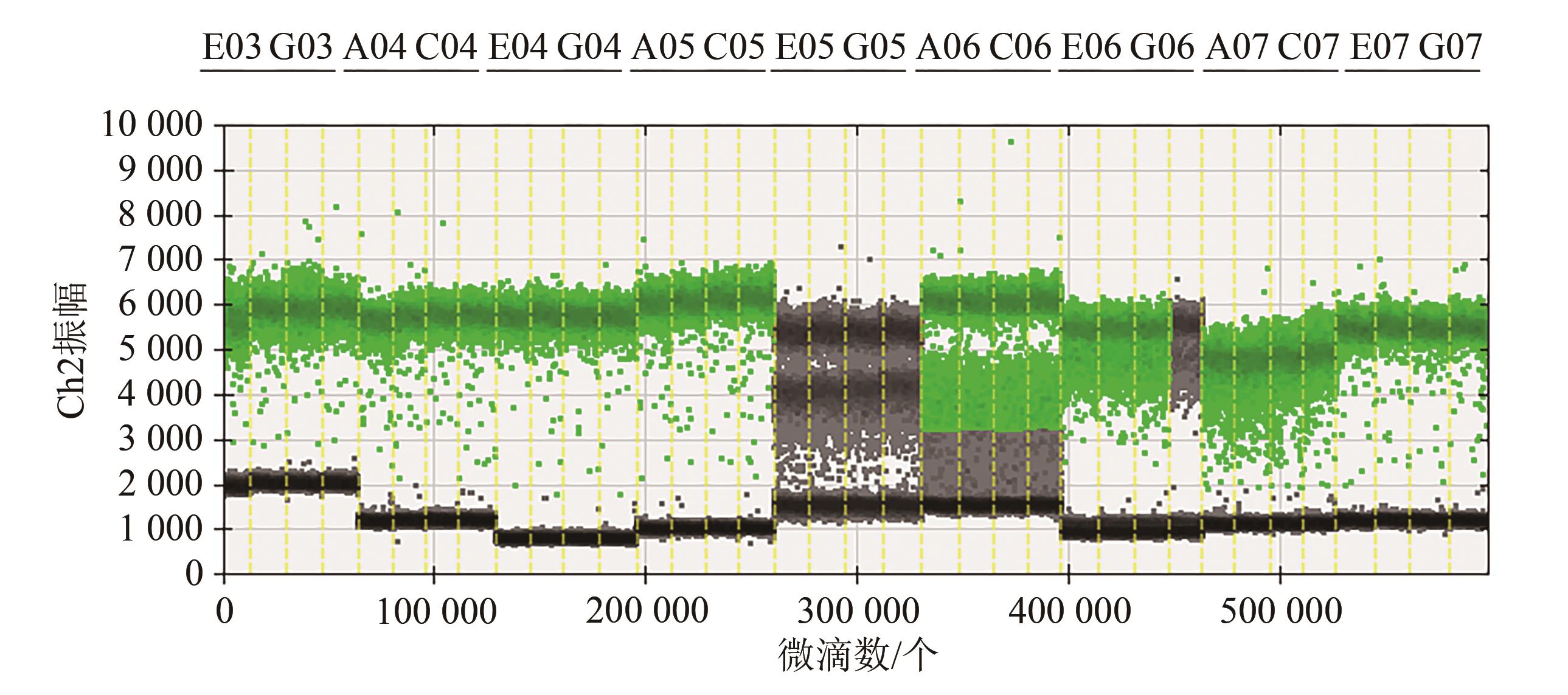
| hmg | 3 550.0 | 3 643.3 | 3 690.0 | 3 653.3 | 3 660.0 | 3 736.7 | 3 655.6±61.78 | 1.69 |
| adh1-1 | 2 630.0 | 2 663.3 | 2 676.7 | 2 676.7 | 2 640.0 | 2 673.3 | 2 660.0±20.22 | 0.76 |
| adh1-2 | 3 190.0 | 3 230.0 | 3 216.7 | 3 260.0 | 3 236.7 | 3 200.0 | 3 222.2±25.53 | 0.79 |
| ivr1-1 | 2 733.3 | 2 753.3 | 2 806.7 | 2 763.3 | 2 813.3 | 2 793.3 | 2 777.2±32.00 | 1.15 |
| ivr1-2 | 3 600.0 | 3 596.7 | 3 790.0 | 3 790.0 | 3 700.0 | 3 716.7 | 3 698.9±86.20 | 2.33 |
| zein-1 | 2 613.3 | 2 666.7 | 2 763.3 | 3 030.0 | 3 583.3 | 4 083.3 | 3 123.3±589.94 | 18.89 |
| zein-2 | 2 823.3 | 2 833.3 | 2 923.3 | 2 900.0 | 2 926.7 | 2 900.0 | 2 884.4±45.00 | 1.56 |
| zSSIIb-1 | 3 443.3 | 3 416.7 | 3 493.3 | 3 496.7 | 3 476.7 | 3 510.0 | 3 472.8±35.80 | 1.03 |
| zSSIIb-2 | 4 070.0 | 4 123.3 | 4 136.7 | 4 186.7 | 4 180.0 | 4 040.0 | 4 122.8±58.63 | 1.42 |
Table 2 Copy number of nine endogenous genes at different annealing temperatures
| 基因名称 | 59.0 ℃ | 58.6 ℃ | 58.0 ℃ | 57.1 ℃ | 55.9 ℃ | 55.0 ℃ | 平均值±标准差 | RSD/% |
|---|---|---|---|---|---|---|---|---|
| hmg | 3 550.0 | 3 643.3 | 3 690.0 | 3 653.3 | 3 660.0 | 3 736.7 | 3 655.6±61.78 | 1.69 |
| adh1-1 | 2 630.0 | 2 663.3 | 2 676.7 | 2 676.7 | 2 640.0 | 2 673.3 | 2 660.0±20.22 | 0.76 |
| adh1-2 | 3 190.0 | 3 230.0 | 3 216.7 | 3 260.0 | 3 236.7 | 3 200.0 | 3 222.2±25.53 | 0.79 |
| ivr1-1 | 2 733.3 | 2 753.3 | 2 806.7 | 2 763.3 | 2 813.3 | 2 793.3 | 2 777.2±32.00 | 1.15 |
| ivr1-2 | 3 600.0 | 3 596.7 | 3 790.0 | 3 790.0 | 3 700.0 | 3 716.7 | 3 698.9±86.20 | 2.33 |
| zein-1 | 2 613.3 | 2 666.7 | 2 763.3 | 3 030.0 | 3 583.3 | 4 083.3 | 3 123.3±589.94 | 18.89 |
| zein-2 | 2 823.3 | 2 833.3 | 2 923.3 | 2 900.0 | 2 926.7 | 2 900.0 | 2 884.4±45.00 | 1.56 |
| zSSIIb-1 | 3 443.3 | 3 416.7 | 3 493.3 | 3 496.7 | 3 476.7 | 3 510.0 | 3 472.8±35.80 | 1.03 |
| zSSIIb-2 | 4 070.0 | 4 123.3 | 4 136.7 | 4 186.7 | 4 180.0 | 4 040.0 | 4 122.8±58.63 | 1.42 |
| MON810稀释倍数 | 拷贝数·µL-1 | 均值 | RSD/% | |||
|---|---|---|---|---|---|---|
| 重复1 | 重复2 | 重复3 | 重复4 | |||
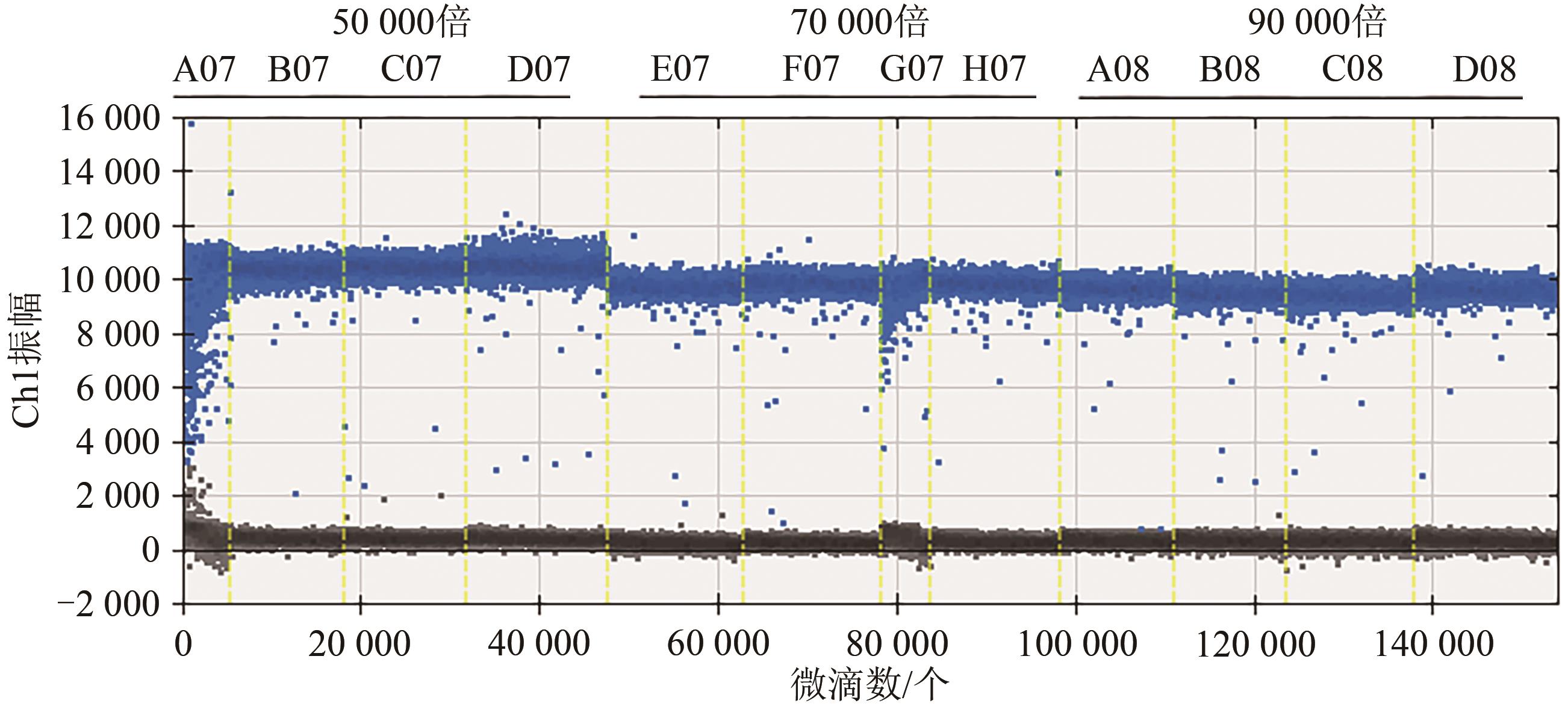
| 50 000 | 5 470 | 5 680 | 5 620 | 5 840 | 5 652.5 | 2.71 |
| 70 000 | 3 680 | 3 760 | 3 600 | 3 880 | 3 730.0 | 3.20 |
| 90 000 | 3 350 | 3 380 | 3 210 | 3 360 | 3 325.0 | 2.34 |
Table 3 Copy number of actual samples MON810
| MON810稀释倍数 | 拷贝数·µL-1 | 均值 | RSD/% | |||
|---|---|---|---|---|---|---|
| 重复1 | 重复2 | 重复3 | 重复4 | |||
| 50 000 | 5 470 | 5 680 | 5 620 | 5 840 | 5 652.5 | 2.71 |
| 70 000 | 3 680 | 3 760 | 3 600 | 3 880 | 3 730.0 | 3.20 |
| 90 000 | 3 350 | 3 380 | 3 210 | 3 360 | 3 325.0 | 2.34 |
| 靶标 | 拷贝数·µL-1 | 均值 | RSD/% | |||
|---|---|---|---|---|---|---|
| 重复1 | 重复2 | 重复3 | 重复4 | |||
| hmg | 3 540 | 3 860 | 3 910 | 3 610 | 3 730.0 | 4.89 |
| adh1-1 | 3 340 | 3 410 | 3 340 | 3 430 | 3 380.0 | 1.39 |
| adh1-2 | 3 920 | 4 030 | 4 000 | 3 970 | 3 980.0 | 1.18 |
| ivr1-1 | 3 260 | 3 400 | 3 440 | 3 420 | 3 380.0 | 2.42 |
| ivr1-2 | No Call | No Call | No Call | No Call | - | - |
| zSSIIb-1 | 3 060 | 3 160 | 3 180 | 3 280 | 3 170.0 | 2.84 |
| zSSIIb-2 | 3 380 | 3 400 | 3 450 | 3 380 | 3 402.5 | 0.97 |
| zein-1 | 4 100 | 4 120 | 4 130 | 4 110 | 4 115.0 | 0.31 |
| zein-2 | 4 120 | 4 130 | 4 120 | 4 040 | 4 102.5 | 1.02 |
Table 4 ddPCR results of nine endogenous genes in mixed DNA solution
| 靶标 | 拷贝数·µL-1 | 均值 | RSD/% | |||
|---|---|---|---|---|---|---|
| 重复1 | 重复2 | 重复3 | 重复4 | |||
| hmg | 3 540 | 3 860 | 3 910 | 3 610 | 3 730.0 | 4.89 |
| adh1-1 | 3 340 | 3 410 | 3 340 | 3 430 | 3 380.0 | 1.39 |
| adh1-2 | 3 920 | 4 030 | 4 000 | 3 970 | 3 980.0 | 1.18 |
| ivr1-1 | 3 260 | 3 400 | 3 440 | 3 420 | 3 380.0 | 2.42 |
| ivr1-2 | No Call | No Call | No Call | No Call | - | - |
| zSSIIb-1 | 3 060 | 3 160 | 3 180 | 3 280 | 3 170.0 | 2.84 |
| zSSIIb-2 | 3 380 | 3 400 | 3 450 | 3 380 | 3 402.5 | 0.97 |
| zein-1 | 4 100 | 4 120 | 4 130 | 4 110 | 4 115.0 | 0.31 |
| zein-2 | 4 120 | 4 130 | 4 120 | 4 040 | 4 102.5 | 1.02 |
| 靶标 | 拷贝数·µL-1 | 均值 | 标准差 | RSD/% | |||
|---|---|---|---|---|---|---|---|
| 重复1 | 重复2 | 重复3 | 重复4 | ||||
| MON810-5 | 547 | 568 | 562 | 584 | 565.25 | 15.31 | 0.03 |
| adh1-1 | 334 | 341 | 334 | 343 | 338.00 | 4.69 | 0.01 |
| 外源/内标 | 164% | 167% | 168% | 170% | 167% | 0.03 | 0.02 |
| MON810-7 | 368 | 376 | 360 | 388 | 373.00 | 11.94 | 0.03 |
| adh1-1 | 334 | 341 | 334 | 343 | 338.00 | 4.69 | 0.01 |
| 外源/内标 | 110% | 110% | 108% | 113% | 110% | 0.02 | 0.02 |
| MON810-9 | 335 | 338 | 321 | 336 | 332.50 | 7.77 | 0.02 |
| adh1-1 | 334 | 341 | 334 | 343 | 338.00 | 4.69 | 0.01 |
| 外源/内标 | 100% | 99% | 96% | 98% | 98% | 0.02 | 0.02 |
Table 5 Comparison result of copy number between MON810 and adh1-1
| 靶标 | 拷贝数·µL-1 | 均值 | 标准差 | RSD/% | |||
|---|---|---|---|---|---|---|---|
| 重复1 | 重复2 | 重复3 | 重复4 | ||||
| MON810-5 | 547 | 568 | 562 | 584 | 565.25 | 15.31 | 0.03 |
| adh1-1 | 334 | 341 | 334 | 343 | 338.00 | 4.69 | 0.01 |
| 外源/内标 | 164% | 167% | 168% | 170% | 167% | 0.03 | 0.02 |
| MON810-7 | 368 | 376 | 360 | 388 | 373.00 | 11.94 | 0.03 |
| adh1-1 | 334 | 341 | 334 | 343 | 338.00 | 4.69 | 0.01 |
| 外源/内标 | 110% | 110% | 108% | 113% | 110% | 0.02 | 0.02 |
| MON810-9 | 335 | 338 | 321 | 336 | 332.50 | 7.77 | 0.02 |
| adh1-1 | 334 | 341 | 334 | 343 | 338.00 | 4.69 | 0.01 |
| 外源/内标 | 100% | 99% | 96% | 98% | 98% | 0.02 | 0.02 |
| 靶标 | 拷贝·µL-1 | 均值 | 标准差 | RSD/% | |||
|---|---|---|---|---|---|---|---|
| 重复1 | 重复2 | 重复3 | 重复4 | ||||
| MON810-5 | 547 | 568 | 562 | 584 | 565.25 | 15.31 | 0.03 |
| adh1-2 | 392 | 403 | 400 | 397 | 398.00 | 4.69 | 0.01 |
| 外源/内标 | 140% | 141% | 141% | 147% | 142% | 0.03 | 0.02 |
| MON810-7 | 368 | 376 | 360 | 388 | 373.00 | 11.94 | 0.03 |
| adh1-2 | 392 | 403 | 400 | 397 | 398.00 | 4.69 | 0.01 |
| 外源/内标 | 94% | 93% | 90% | 98% | 94% | 0.03 | 0.03 |
| MON810-9 | 335 | 338 | 321 | 336 | 332.50 | 7.77 | 0.02 |
| adh1-2 | 392 | 403 | 400 | 397 | 398.00 | 4.69 | 0.01 |
| 外源/内标 | 85% | 84% | 80% | 85% | 84% | 0.02 | 0.03 |
Table 6 Comparison result of copy number between MON810 and adh1-2
| 靶标 | 拷贝·µL-1 | 均值 | 标准差 | RSD/% | |||
|---|---|---|---|---|---|---|---|
| 重复1 | 重复2 | 重复3 | 重复4 | ||||
| MON810-5 | 547 | 568 | 562 | 584 | 565.25 | 15.31 | 0.03 |
| adh1-2 | 392 | 403 | 400 | 397 | 398.00 | 4.69 | 0.01 |
| 外源/内标 | 140% | 141% | 141% | 147% | 142% | 0.03 | 0.02 |
| MON810-7 | 368 | 376 | 360 | 388 | 373.00 | 11.94 | 0.03 |
| adh1-2 | 392 | 403 | 400 | 397 | 398.00 | 4.69 | 0.01 |
| 外源/内标 | 94% | 93% | 90% | 98% | 94% | 0.03 | 0.03 |
| MON810-9 | 335 | 338 | 321 | 336 | 332.50 | 7.77 | 0.02 |
| adh1-2 | 392 | 403 | 400 | 397 | 398.00 | 4.69 | 0.01 |
| 外源/内标 | 85% | 84% | 80% | 85% | 84% | 0.02 | 0.03 |
| 1 | 王凤军,陈强,叶素丹,等.转基因玉米品系及转化体成分四重实时荧光PCR快速鉴定[J].中国粮油学报,2021,36(12):128-135. |
| WANG F J, CHEN Q, YE S D, et al.. Rapid identification of transgenic maize lines and transformants by quadruplex real-time PCR[J]. J. Chin. Cereals Oils Assoc., 2021, 36(12): 128-135. | |
| 2 | TAKABATAKE R, ONISHI M, FUTO S, et al.. Comparison of the specificity,stability,and PCR efficiency of six rice endogenous sequences for detection analyses of genetically modified rice[J]. Food Contr., 2015, 50: 949-955. |
| 3 | 陈利红,周俊飞,梁晋刚,等.基于二代测序技术的玉米内标准基因扩增子的筛选与评估[J].食品科学,2023,44(20):146-154. |
| CHEN L H, ZHOU J F, LIANG J G, et al.. Screening and evaluation of amplicons of maize endogenous reference genes based on next-generation sequencing technology[J]. Food Sci., 2023, 44(20): 146-154. | |
| 4 | 高佳奇,陈硕,王迪 等.六种作物内标准基因开发与检测研究进展[J]. 生物技术进展, 2020, 10(6): 613-622. |
| GAO J Q, CHEN S, WANG D, et al.. Progress on development and detection of internal reference genes in six crops[J]. Curr. Biotechnol., 2020, 10(6): 613-622. | |
| 5 | COTTENET G, BLANCPAIN C, CHUAH P F. Performance assessment of digital PCR for the quantification of GM-maize and GM-soya events[J]. Anal. Bioanal. Chem., 2019, 411(11): 2461-2469. |
| 6 | DENG T T, HUANG W S, REN J N, et al.. Verification and applicability of endogenous reference genes for quantifying GM rice by digital PCR[J/OL]. Anal. Biochem., 2019, 587: 113442[2024-12-30]. . |
| 7 | PATERNÒ A, MARCHESI U, GATTO F, et al.. Finding the joker among the maize endogenous reference genes for genetically modified organism (GMO) detection[J]. J. Agric. Food Chem., 2009, 57(23): 11086-11091. |
| 8 | 王凤军,杨伊平,叶素丹,等.多重荧光定量PCR快速鉴定转基因玉米品系[J].中国粮油学报,2023,38(4):143-149. |
| WANG F J, YANG Y P, YE S D, et al.. Rapid identification of transgenic maize lines by multiplex fluorescence quantitative PCR[J]. J. Chin. Cereals Oils Assoc., 2023, 38(4): 143-149. | |
| 9 | 李凌燕,张旭冬,陈子言,等.转基因抗虫耐除草剂玉米MON87411精准定量检测方法的建立[J].生物安全学报,2023,32(1):38-45. |
| LI L Y, ZHANG X D, CHEN Z Y, et al.. Establishment of accurate quantitative detection method for insect-resistant and herbicide-tolerant maize MON87411[J]. J. Biosaf., 2023, 32(1): 38-45. | |
| 10 | 周圆,单长林,李孝军,等.运用微滴式数字PCR技术检测转基因玉米品系的研究[J].安徽农业科学,2018,46(14):175-178. |
| ZHOU Y, SHAN C L, LI X J, et al.. Study on detection of genetically modified maize lines using microdrop digital PCR technique[J]. J. Anhui Agric. Sci., 2018, 46(14): 175-178. | |
| 11 | FOLLONI S, BELLOCCHI G, PROSPERO A, et al.. Statistical evaluation of real-time PCR protocols applied to quantify genetically modified maize[J]. Food Anal. Meth., 2010, 3(4): 304-312. |
| 12 | WEI S, WANG C G, ZHU P Y, et al.. A high-throughput multiplex tandem PCR assay for the screening of genetically modified maize[J]. LWT, 2018, 87: 169-176. |
| 13 | PAPAZOVA N, ZHANG D, GRUDEN K, et al.. Evaluation of the reliability of maize reference assays for GMO quantification[J]. Anal. Bioanal. Chem., 2010, 396(6): 2189-2201. |
| 14 | 肖芳,李俊,王颢潜 等.转基因玉米NK603转化体/zSSIIb内标基因二重微滴数字PCR方法的建立及应用[J]. 中国农业科学, 2021, 54(22): 4728-4739. |
| XIAO F, LI J, WANG H Q, et al.. Establishment and application of A duplex ddPCR method to quantify the NK603/zSSIIb copy number ratio in transgenic maize NK603[J]. Sci. Agric. Sin., 2021, 54(22): 4728-4739. | |
| 15 | SCHOLDBERG T A, NORDEN T D, NELSON D D, et al.. Evaluating precision and accuracy when quantifying different endogenous control reference genes in maize using real-time PCR[J]. J. Agric. Food Chem., 2009, 57(7): 2903-2911. |
| 16 | DMIQE G, HUGGETT J F.The digital MIQE guidelines update:minimum information for publication of quantitative digital PCR experiments for 2020[J]. Clin. Chem., 2020, 66(8): 1012-1029. |
| 17 | BASU A S. Digital assays part I: partitioning statistics and digital PCR[J]. SLAS Technol., 2017, 22(4): 369-386. |
| 18 | CORBISIER P, BUTTINGER G, SAVINI C, et al.. Expression of GM content in mass fraction from digital PCR data[J/OL]. Food Contr., 2022, 133(Pt B): 108626[2024-12-30]. . |
| 19 | ROWLANDS V, RUTKOWSKI A J, MEUSER E, et al.. Optimisation of robust singleplex and multiplex droplet digital PCR assays for high confidence mutation detection in circulating tumour DNA[J/OL]. Sci. Rep., 2019, 9: 12620[2024-12-30]. . |
| 20 | ZHANG H, LAŠŠÁKOVÁ S, YAN Z, et al.. Digital polymerase chain reaction duplexing method in a single fluorescence channel[J/OL]. Anal. Chim. Acta., 2023, 1238: 340243[2024-12-30]. . |
| 21 | DEBSKI P R, GARSTECKI P. Designing and interpretation of digital assays: concentration of target in the sample and in the source of sample[J]. Biomol. Detect. Quantif., 2016, 10: 24-30. |
| 22 | 国家质量监督检验检疫总局,中国国家标准化管理委员会. 水溶液中核酸的浓度和纯度检测 紫外分光光度法: [S].北京:中国标准出版社,2017. |
| 23 | 刘晓,朱鹏宇,景小艳,等.双重数字PCR在转基因大豆检测中的应用[J].生物技术进展,2020,10(1):60-66. |
| LIU X, ZHU P Y, JING X Y, et al.. Application of duplex droplet digital PCR for detection of genetically modified soybean[J]. Curr. Biotechnol., 2020, 10(1): 60-66. | |
| 24 | 卢海强,焦新雅,吴思源,等.数字PCR在食源性致病菌检测中的应用进展[J]. 生物技术进展, 2021, 11(3): 260-268. |
| LU H Q, JIAO X Y, WU S Y, et al.. Application progress on the digital PCR in detection of foodborne pathogenic bacteria[J]. Curr. Biotechnol., 2021, 11(3): 260-268. |
| [1] | Yanyan JIA, Luyang DUANMU. Study on the Mechanism of Bubble Generation and Inhibition Method During Digital PCR Amplification Process [J]. Current Biotechnology, 2025, 15(4): 693-701. |
| [2] | Hongbo FAN, Liangyong HU, Songqing HU. Establishment of Digital PCR Detection System for Helicobacter pyloriureC and 23S rDNA [J]. Current Biotechnology, 2024, 14(5): 868-874. |
| [3] | Yi JI, Kaili WANG, Huiru YU, Xin ZHAO, Lin DING, Cheng PENG, Junfeng XU, Xiaoyun CHEN. Development of Sheep-derived Genomic DNA Reference Material [J]. Current Biotechnology, 2024, 14(1): 125-132. |
| [4] | Peixuan MEN, Yufeng XIAO, Bin ZHANG. Analysis of Clinical Application and Technology Hotspots of Digital PCR Technology Based on Bibliometrics Methods [J]. Current Biotechnology, 2022, 12(4): 606-613. |
| [5] | Xing DANG, Binwei ZHI, Kehao CAO, Tingting LIU, Biao CHEN, Yuanjie DING. Patent Analysis on Genetically Modified Maize Biological Breeding Technology and Development Suggestions [J]. Current Biotechnology, 2022, 12(4): 614-622. |
| [6] | Hongtao WEN, Yang YANG, Yijia DING, Ran YUAN, Ruiying ZHANG, Zhiyuan XU, Jingang LIANG. Establishment of Qualitative PCR Detection of Transgenic Insect⁃resistant Maize CM8101 [J]. Current Biotechnology, 2022, 12(2): 248-255. |
| [7] | LU Haiqiang1, JIAO Xinya1,2, WU Siyuan2, WANG Yali2, LIU Lu2, CHENG Shumei1, ZHANG Xiao3*, SU Xiaofeng2*. Application Progress on the Digital PCR in Detection of Foodborne Pathogenic Bacteria [J]. Curr. Biotech., 2021, 11(3): 260-268. |
| [8] | WANG Di, WU Xiao, WANG Zhidong, GAO Yunhua*. Research Progress on the Flow Cytometry Counting for Single Nucleotide Molecule [J]. Curr. Biotech., 2020, 10(6): 573-578. |
| [9] | ZHENG Zifan, LIU Fangfang, LIU Weixiao, JIN Wujun, LI Liang*. Application of Digital PCR Technology in the Development of Nucleic Acid Reference Materials [J]. Curr. Biotech., 2020, 10(6): 579-584. |
| [10] | WANG Shangjun1, DONG Lianhua2. Research on Calibration Method of Digital PCR Instrument [J]. Curr. Biotech., 2020, 10(6): 585-589. |
| [11] | HU Sihong, YOU Guoye. Application Prospects of Digital PCR in Detection of SARS-CoV-2 [J]. Curr. Biotech., 2020, 10(6): 674-679. |
| [12] | REN Wen1, YANG Haixia2, CHEN Lizhu2, LI Yufeng2, LIU Ya1*. Establishment and Application of Nucleic Acid Chromatography for Rapid Detection of Transgenic Plants [J]. Curr. Biotech., 2020, 10(6): 680-687. |
| [13] | WEN Hongtao1§,YANG Yang1§,DING Yijia1,YUAN Ran1,ZHANG Xiujie2,ZHANG Ruiying1*. Qualitative PCR Methods for the Detection of Transgenic Herbicide-tolerant Maize G1105E-823C [J]. Curr. Biotech., 2020, 10(6): 688-695. |
| [14] | JI Yi, CHEN Xiaoyun, DING Lin, WANG Xiaofu, PENG Cheng, XU Xiaoli, XU Junfeng*. Progress on Identification Methods of Animal-derived Ingredients in Meat and Meat Products [J]. Curr. Biotech., 2020, 10(6): 711-716. |
| [15] | WU Wenyan§, LIU Xinxiang§, ZHOU Miaoyi*, LIU Jinxiang, LIU Ya. Establishment of Event-specific PCR Detection Method of Transgenic Maize Line 2A-5 [J]. Curr. Biotech., 2020, 10(4): 363-370. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||