Current Biotechnology ›› 2024, Vol. 14 ›› Issue (1): 125-132.DOI: 10.19586/j.2095-2341.2023.0137
• Articles • Previous Articles Next Articles
Development of Sheep-derived Genomic DNA Reference Material
Yi JI1( ), Kaili WANG2, Huiru YU3, Xin ZHAO4, Lin DING1, Cheng PENG1, Junfeng XU1, Xiaoyun CHEN1(
), Kaili WANG2, Huiru YU3, Xin ZHAO4, Lin DING1, Cheng PENG1, Junfeng XU1, Xiaoyun CHEN1( )
)
- 1.State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products,Key Laboratory of Traceability for Agricultural Genetically Modified Organisms,Ministry of Agriculture and Rural Affairs,Zhejiang Academy of Agricultural Sciences,Hangzhou 310021,China
2.School of Food and Pharmacy,Ningbo University,Zhejiang Ningbo 215211,China
3.Xianghu Laboratory,Hangzhou 311231,China
4.Institute of Germplasm Resources and Biotechnology,Tianjin Academy of Agricultural Sciences,Tianjin 300384,China
-
Received:2023-10-25Accepted:2023-11-25Online:2024-01-25Published:2024-02-05 -
Contact:Xiaoyun CHEN
羊源性基因组DNA标准物质研制
纪艺1( ), 王凯莉2, 余卉茹3, 赵新4, 丁霖1, 彭城1, 徐俊锋1, 陈笑芸1(
), 王凯莉2, 余卉茹3, 赵新4, 丁霖1, 彭城1, 徐俊锋1, 陈笑芸1( )
)
- 1.浙江省农业科学院,农产品质量安全危害因子与风险防控国家重点实验室,农业农村部农业转基因生物溯源重点实验室,杭州 310021
2.宁波大学食品与药学学院,浙江 宁波 215211
3.湘湖实验室,杭州 311231
4.天津市农业科学院,种质资源与生物技术研究所,天津 300384
-
通讯作者:陈笑芸 -
作者简介:纪艺E-mail: jymemory12138@163.com; -
基金资助:科技创新专项2030重大项目农业生物育种重大专项(2022ZD0402012);浙江省重点研发项目(2021C02059)
CLC Number:
Cite this article
Yi JI, Kaili WANG, Huiru YU, Xin ZHAO, Lin DING, Cheng PENG, Junfeng XU, Xiaoyun CHEN. Development of Sheep-derived Genomic DNA Reference Material[J]. Current Biotechnology, 2024, 14(1): 125-132.
纪艺, 王凯莉, 余卉茹, 赵新, 丁霖, 彭城, 徐俊锋, 陈笑芸. 羊源性基因组DNA标准物质研制[J]. 生物技术进展, 2024, 14(1): 125-132.
share this article
| 标准物质 | 组间自由度 | 组内自由度 | 组间均方 | 组内均方 | 统计量(F) | F0.05(5,12) |
|---|---|---|---|---|---|---|
| HELZ(高浓度) | 5 | 12 | 1.48×104 | 9.90×103 | 1.49 | 3.11 |
| HELZ(低浓度) | 5 | 12 | 1.41×102 | 1.29×102 | 1.09 | 3.11 |
Table 1 Results of statistical analysis of the uniformity
| 标准物质 | 组间自由度 | 组内自由度 | 组间均方 | 组内均方 | 统计量(F) | F0.05(5,12) |
|---|---|---|---|---|---|---|
| HELZ(高浓度) | 5 | 12 | 1.48×104 | 9.90×103 | 1.49 | 3.11 |
| HELZ(低浓度) | 5 | 12 | 1.41×102 | 1.29×102 | 1.09 | 3.11 |
| 标准物质 | 组间自由度 | 组内自由度 | 组间均方 | 组内均方 | F0.05(11,24) | ||
|---|---|---|---|---|---|---|---|
| HELZ(高浓度) | 11 | 24 | 1.05×104 | 1.03×102 | 54.23 | 1.1% | 2.215 |
| HELZ(低浓度) | 11 | 24 | 2.96×102 | 2.06×101 | 7.96 | 1.5% | 2.215 |
Table 2 Uniformity statistical analysis results
| 标准物质 | 组间自由度 | 组内自由度 | 组间均方 | 组内均方 | F0.05(11,24) | ||
|---|---|---|---|---|---|---|---|
| HELZ(高浓度) | 11 | 24 | 1.05×104 | 1.03×102 | 54.23 | 1.1% | 2.215 |
| HELZ(低浓度) | 11 | 24 | 2.96×102 | 2.06×101 | 7.96 | 1.5% | 2.215 |
| 标准物质 | HELZ(高浓度) | HELZ(低浓度) | ||
|---|---|---|---|---|
| 4 ℃ | 25 ℃ | 4 ℃ | 25 ℃ | |
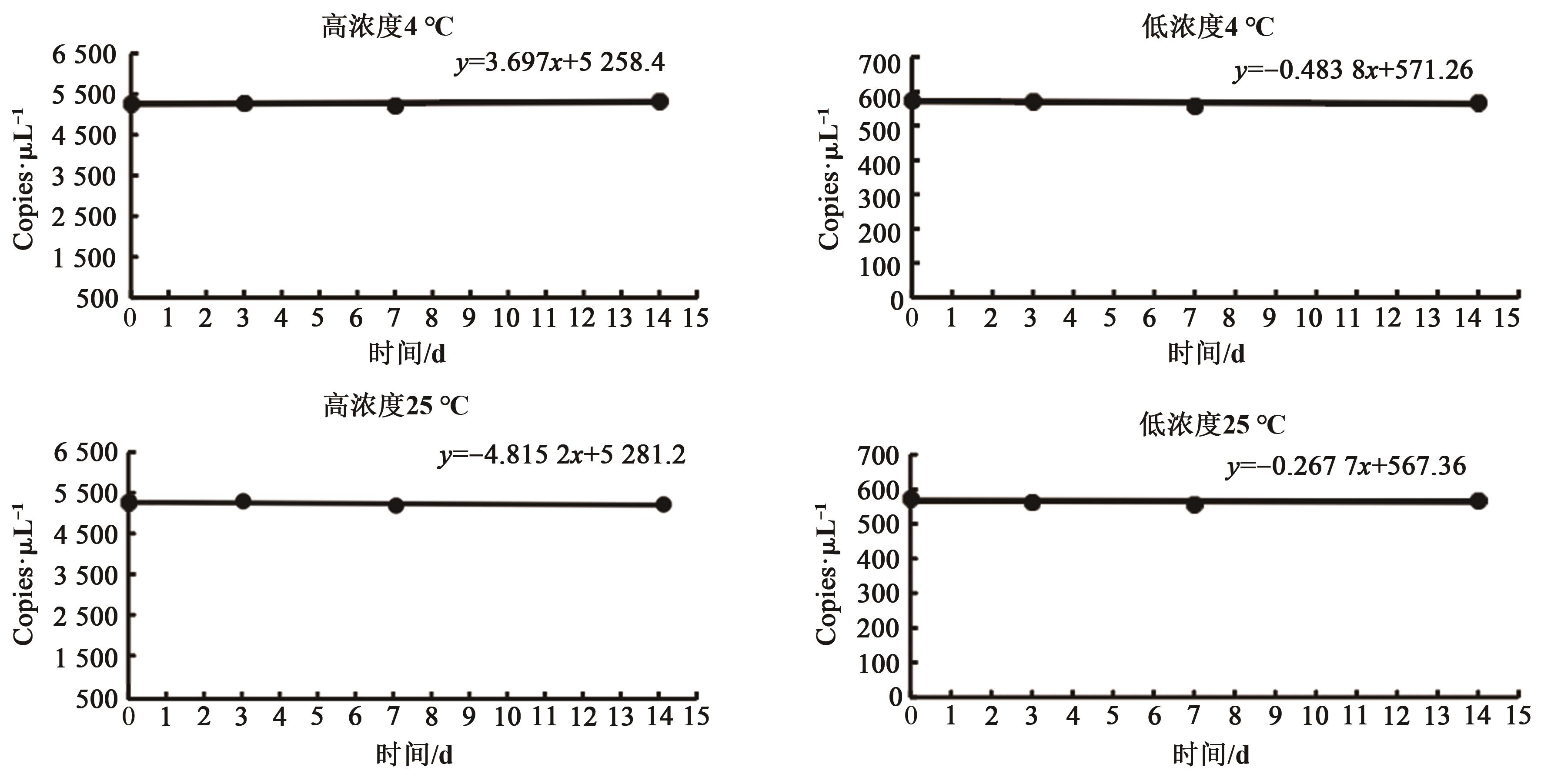
| 总均值 | 5.28×103 | 5.25×103 | 5.68×102 | 5.66×102 |
| β1 | 3.70 | -4.82 | -0.48 | -0.27 |
| 截距β0 | 5 258.37 | 5 281.60 | 571.26 | 567.36 |
| 标准偏差S(β1) | 4.42 | 4.48 | 0.76 | 0.81 |
| t0.95,2 | 4.3 | 4.3 | 4.3 | 4.3 |
| t0.95,2×S(β1) | 19.006 | 19.264 | 3.268 | 3.483 |
| 稳定性判断 | │β1│<t0.95,3×S(β1) | |||
| 判断结果 | 稳定 | 稳定 | 稳定 | 稳定 |
Table 3 Short-term stability assessment results
| 标准物质 | HELZ(高浓度) | HELZ(低浓度) | ||
|---|---|---|---|---|
| 4 ℃ | 25 ℃ | 4 ℃ | 25 ℃ | |
| 总均值 | 5.28×103 | 5.25×103 | 5.68×102 | 5.66×102 |
| β1 | 3.70 | -4.82 | -0.48 | -0.27 |
| 截距β0 | 5 258.37 | 5 281.60 | 571.26 | 567.36 |
| 标准偏差S(β1) | 4.42 | 4.48 | 0.76 | 0.81 |
| t0.95,2 | 4.3 | 4.3 | 4.3 | 4.3 |
| t0.95,2×S(β1) | 19.006 | 19.264 | 3.268 | 3.483 |
| 稳定性判断 | │β1│<t0.95,3×S(β1) | |||
| 判断结果 | 稳定 | 稳定 | 稳定 | 稳定 |
| 标准物质 | HELZ(高浓度) | HELZ(低浓度) |
|---|---|---|
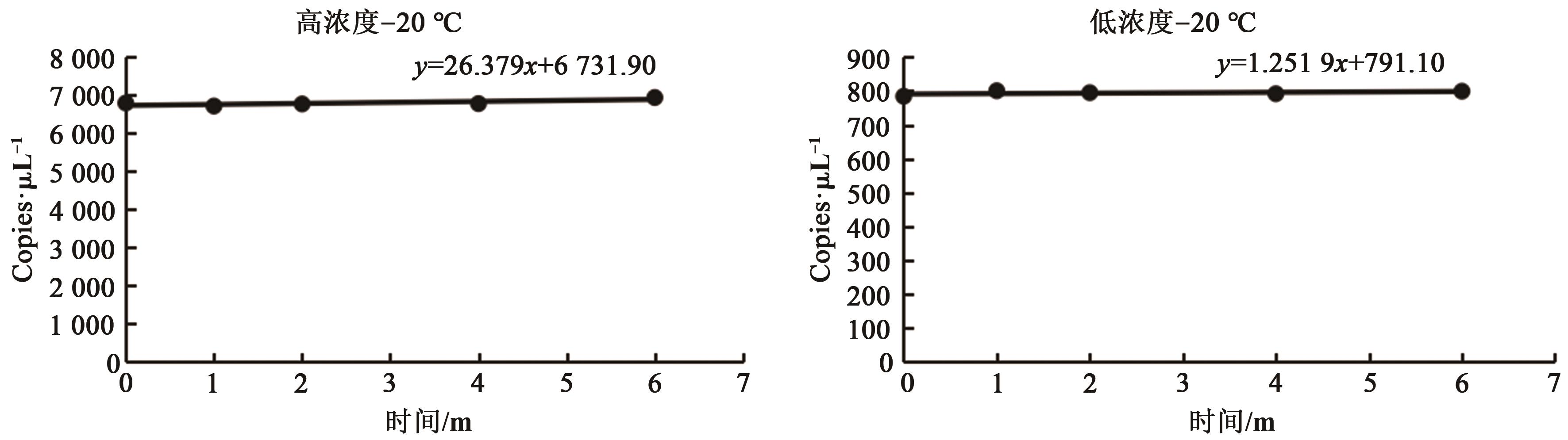
| 均值 | 5.21×103 | 5.43×102 |
| β1 | 7.92 | 0.32 |
| 截距β0 | 5 192.56 | 562.72 |
| 标准偏差S(β1) | 15.07 | 1.61 |
| t0.95,3 | 3.18 | 3.18 |
| t0.95,3×S(β1) | 47.90 | 5.12 |
| 稳定性判断 | │β1│<t0.95,3×S(β1) | |
| 判断结果 | 稳定 | 稳定 |
Table 4 Evaluation results of long-term stability at -20 ℃
| 标准物质 | HELZ(高浓度) | HELZ(低浓度) |
|---|---|---|
| 均值 | 5.21×103 | 5.43×102 |
| β1 | 7.92 | 0.32 |
| 截距β0 | 5 192.56 | 562.72 |
| 标准偏差S(β1) | 15.07 | 1.61 |
| t0.95,3 | 3.18 | 3.18 |
| t0.95,3×S(β1) | 47.90 | 5.12 |
| 稳定性判断 | │β1│<t0.95,3×S(β1) | |
| 判断结果 | 稳定 | 稳定 |
| 标准物质 | HELZ(高浓度) | HELZ(低浓度) |
|---|---|---|
| 均值 | 5.24×103 | 5.69×102 |
| β1 | 9.37 | -0.40 |
| 截距β0 | 5 183.78 | 570.80 |
| 标准偏差S(β1) | 5.06 | 0.93 |
| t0.95,8 | 2.31 | 2.31 |
| tβ1 | 1.85 | 0.43 |
| 稳定性判断 | tβ1<t0.95,8 | |
| 判断结果 | 稳定 | 稳定 |
Table 5 Evaluation results of freeze-thaw stability
| 标准物质 | HELZ(高浓度) | HELZ(低浓度) |
|---|---|---|
| 均值 | 5.24×103 | 5.69×102 |
| β1 | 9.37 | -0.40 |
| 截距β0 | 5 183.78 | 570.80 |
| 标准偏差S(β1) | 5.06 | 0.93 |
| t0.95,8 | 2.31 | 2.31 |
| tβ1 | 1.85 | 0.43 |
| 稳定性判断 | tβ1<t0.95,8 | |
| 判断结果 | 稳定 | 稳定 |
| 实验室编号 | Copies·μL-1 | |
|---|---|---|
| HELZ(高浓度) | HELZ(低浓度) | |
| 1 | 5.18×103 | 5.40×102 |
| 2 | 5.59×103 | 5.83×102 |
| 3 | 5.71×103 | 5.87×102 |
| 4 | 5.45×103 | 5.55×102 |
| 5 | 5.52×103 | 5.78×102 |
| 6 | 5.33×103 | 5.69×102 |
| 7 | 5.41×103 | 5.56×102 |
| 8 | 5.35×103 | 5.74×102 |
| 9 | 5.41×103 | 5.68×102 |
| 总平均值 | 5.44×103 | 5.68×102 |
| SD | 1.55×102 | 1.51×101 |
| RSD/% | 2.85 | 2.66 |
Table 6 Analysis of joint setting data from 9 laboratories
| 实验室编号 | Copies·μL-1 | |
|---|---|---|
| HELZ(高浓度) | HELZ(低浓度) | |
| 1 | 5.18×103 | 5.40×102 |
| 2 | 5.59×103 | 5.83×102 |
| 3 | 5.71×103 | 5.87×102 |
| 4 | 5.45×103 | 5.55×102 |
| 5 | 5.52×103 | 5.78×102 |
| 6 | 5.33×103 | 5.69×102 |
| 7 | 5.41×103 | 5.56×102 |
| 8 | 5.35×103 | 5.74×102 |
| 9 | 5.41×103 | 5.68×102 |
| 总平均值 | 5.44×103 | 5.68×102 |
| SD | 1.55×102 | 1.51×101 |
| RSD/% | 2.85 | 2.66 |
| 标准物质 | 标准值Y | 定值相对不确定度 | 均匀性相对不确定度 | 短期稳定性相对不确定度 | 长期稳定性相对不确定度 | 相对标准不确定度 | 相对扩展不确定度 | 扩展不确定度UCRM (k=2)/(copies·μL-1) |
|---|---|---|---|---|---|---|---|---|
| HELZ(高浓度) | 5.44×103 | 0.033 | 0.011 | 0.012 | 0.018 | 0.041 | 0.082 | 0.45×103 |
| HELZ(低浓度) | 5.68×102 | 0.035 | 0.015 | 0.020 | 0.018 | 0.047 | 0.094 | 0.54×102 |
Table 7 Uncertainty evaluation results
| 标准物质 | 标准值Y | 定值相对不确定度 | 均匀性相对不确定度 | 短期稳定性相对不确定度 | 长期稳定性相对不确定度 | 相对标准不确定度 | 相对扩展不确定度 | 扩展不确定度UCRM (k=2)/(copies·μL-1) |
|---|---|---|---|---|---|---|---|---|
| HELZ(高浓度) | 5.44×103 | 0.033 | 0.011 | 0.012 | 0.018 | 0.041 | 0.082 | 0.45×103 |
| HELZ(低浓度) | 5.68×102 | 0.035 | 0.015 | 0.020 | 0.018 | 0.047 | 0.094 | 0.54×102 |
| 1 | SALTER A M. The effects of meat consumption on global health[J]. Rev. Sci. Tech., 2018, 37(1): 47-55. |
| 2 | KÖPPEL R, DANIELS M, FELDERER N, et al.. Multiplex real-time PCR for the detection and quantification of DNA from duck, goose, chicken, turkey and pork[J]. Eur. Food Res. Technol., 2013, 236(6): 1093-1098. |
| 3 | 石盼盼,李旭,魏法山,等.肉类掺假的分子生物学检测[J].食品与生物技术学报,2017,36(7):773-777. |
| SHI P P, LI X, WEI F S, et al.. Molecular biological detection for adulterated meat[J]. J. Food Sci. Biotechnol., 2017, 36(7): 773-777. | |
| 4 | 李宗梦,赵良娟,赵宏,等.肉及肉制品动物源性成分鉴别技术研究进展[J].食品研究与开发,2014,35(18):122-126+127. |
| LI Z M, ZHAO L J, ZHAO H, et al.. Research progress in identification techniques of animal ingredient in meat and meat products[J]. Food Res. Dev., 2014, 35(18): 122-126+127. | |
| 5 | KUMAR A, KUMAR R R, SHARMA B D, et al.. Identification of species origin of meat and meat products on the DNA basis: a review[J]. Crit. Rev. Food Sci. Nutr., 2015, 55(10): 1340-1351. |
| 6 | CAVIN C, COTTENET G, COOPER K M, et al.. Meat vulnerabilities to economic food adulteration require new analytical solutions[J]. Chim. Aarau., 2018, 72(10): 697-703. |
| 7 | ZIA Q, ALAWAMI M, MOKHTAR N F K, et al.. Current analytical methods for porcine identification in meat and meat products[J/OL]. Food Chem., 2020, 324: 126664[2023-10-12]. . |
| 8 | STACHNIUK A, SUMARA A, MONTOWSKA M, et al.. Liquid chromatography-mass spectrometry bottom-up proteomic methods in animal species analysis of processed meat for food authentication and the detection of adulterations[J]. Mass Spectrom. Rev., 2021, 40(1): 3-30. |
| 9 | KITPIPIT T, SITTICHAN K, THANAKIATKRAI P. Are these food products fraudulent? Rapid and novel triplex-direct PCR assay for meat identification[J]. Foren. Sci. Intern.Genet. , 2013, 4(1): 33-34. |
| 10 | XU R, WEI S, ZHOU G, et al.. Multiplex TaqMan locked nucleic acid real-time PCR for the differential identification of various meat and meat products[J]. Meat Sci., 2018, 137: 41-46. |
| 11 | LI T, WANG J, WANG Z, et al.. Quantitative determination of mutton adulteration with single-copy nuclear genes by real-time PCR[J/OL]. Food Chem., 2021, 344: 128622[2023-12-01]. . |
| 12 | FLOREN C, WIEDEMANN I, BRENIG B, et al.. Species identification and quantification in meat and meat products using droplet digital PCR (ddPCR)[J]. Food Chem., 2015, 173: 1054-1058. |
| 13 | LIN L, ZHENG Y, HUANG H, et al.. A visual method to detect meat adulteration by recombinase polymerase amplification combined with lateral flow dipstick[J/OL]. Food Chem., 2021, 354: 129526[2023-10-11]. . |
| 14 | WANG J, WAN Y, CHEN G, et al.. Colorimetric detection of horse meat based on loop-mediated isothermal amplification (LAMP)[J]. Food Anal. Meth., 2019, 12(11): 2535-2541. |
| 15 | CAO Y, ZHENG K, JIANG J, et al.. A novel method to detect meat adulteration by recombinase polymerase amplification and SYBR green I[J]. Food Chem., 2018, 266: 73-78. |
| 16 | HOSSAIN M A, ALI M E, HAMID S B ABD, et al.. Double gene targeting multiplex polymerase chain reaction–restriction fragment length polymorphism assay discriminates beef, buffalo, and pork substitution in frankfurter products[J]. J. Agric. Food Chem., 2016, 64(32): 6343-6354. |
| 17 | LOPEZ-OCEJA A, NUÑEZ C, BAETA M, et al.. Species identification in meat products: a new screening method based on high resolution melting analysis of cyt b gene[J]. Food Chem., 2017, 237: 701-706. |
| 18 | 叶德培,施昌彦,金华彰,等.通用计量术语及定义 JJF 1001-2011[M].北京: 中国标准出版社. |
| 19 | QIN Y, ZHANG X, DAI X, et al.. Graphene oxide-assisted synthesis of Pt-co alloy nanocrystals with high-index facets and enhanced electrocatalytic properties[J]. Small, 2016, 12(4): 524-533. |
| 20 | YANG Y, LI L, YANG H, et al.. Development of certified matrix-based reference material as a calibrator for genetically modified rice G6H1 analysis[J]. J. Agric. Food Chem., 2018, 66(14): 3708-3715. |
| 21 | LI J, LI L, ZHANG L, et al.. Development of a certified genomic DNA reference material for detection and quantification of genetically modified rice KMD[J]. Anal. Bioanal. Chem., 2020, 412(25): 7007-7016. |
| 22 | XU J, QU S, SUN N, et al.. Construction of a reference material panel for detecting KRAS/NRAS/EGFR/BRAF/MET mutations in plasma ctDNA[J]. J. Clin. Pathol., 2021, 74(5): 314-320. |
| 23 | DONG L, WANG X, WANG S, et al.. Interlaboratory assessment of droplet digital PCR for quantification of BRAF V600E mutation using a novel DNA reference material[J/OL]. Talanta, 2020, 207: 120293[2023-10-12]. . |
| 24 | HE H J, DAS B, CLEVELAND M H, et al.. Development and interlaboratory evaluation of a NIST Reference Material RM 8366 for EGFR and MET gene copy number measurements[J]. Clin. Chem. Lab. Med., 2019, 57(8): 1142-1152. |
| 25 | VALLEJO C V, TERE C P, CALDERON M N, et al.. Development of a genomic DNA reference material for Salmonella enteritidis detection using polymerase chain reaction[J/OL]. Mol. Cell Probes, 2021, 55: 101690[2023-10-13]. . |
| 26 | STEFFEN C R, KIESLER K M, BORSUK L A, et al.. Beyond the STRs: a comprehensive view of current forensic DNA markers characterized in the PCR-based DNA profiling standard SRM 2391D[J/OL]. Foren. Sci. Int. Genet. Suppl. Series, 2017,6:426-427. |
| 27 | QIONG H E, CHANGHUI L, HUIJUN W, et al.. The method development of typing multiplex-SNP with fluorescein-labeled and investigation of SNP-pseudohaplotype of mitochondrial DNA[J]. Acta Metall. Sinica, 2011(5):81-82. |
| 28 | ZHAO S, ZHAO Y. Application and preparation progress of stable isotope reference materials in traceability of agricultural products[J]. Crit. Rev. Anal. Chem., 2021, 51(8): 742-753. |
| 29 | 卢晓华,汪斌,周桃庚,等.标准物质的定值及均匀性、稳定性评估 JJF 1343-2022[M]. 北京: 中国标准出版社, 2022: 1-66. |
| 30 | 游英华,赵志,李亚楠,等.数字PCR在转基因植物检测中的应用[J].中国生物防治学报,2022,38(5):1143-1148. |
| YOU Y H, ZHAO Z, LI Y N, et al.. Application of digital PCR in the detection of genetically modified plants[J]. Chin. J. Biol. Contr., 2022, 38(5): 1143-1148. | |
| 31 | 陈林军,崔强,于志超,等.数字PCR技术在动物疫病检测中的应用[J].畜牧与饲料科学,2021,42(6):124-128. |
| CHEN L J, CUI Q, YU Z C, et al.. Application of digital PCR assay in detection of animal epidemic diseases[J]. Anim. Husb. Feed. Sci., 2021, 42(6): 124-128. | |
| 32 | 王森,陈东亮,黄丛林,等.基于PCR的植物病毒检测技术研究进展[J].植物检疫,2021,35(3):1-8. |
| WANG S, CHEN D L, HUANG C L, et al.. Progress in PCR-based techniques for detection of plant viruses[J]. Plant Quar., 2021, 35(3): 1-8. | |
| 33 | 王霞,郭若晖,董莲华.新型冠状病毒B.1.1.7变异株特异性数字PCR定量方法研究[J].计量学报,2023(5):826-832. |
| WANG X, GUO R H, DONG L H. The establishment of digital PCR quantitative method for strain B.1.1.7 of SARS-CoV-2[J]. Acta Metrol. Sin., 2023(5): 826-832. | |
| 34 | 门佩璇,肖宇锋,张玢.基于计量学方法分析数字PCR技术的临床应用现状与技术热点[J].生物技术进展,2022,12(4):606-613. |
| MEN P X, XIAO Y F, ZHANG F. Analysis of clinical application and technology hotspots of digital PCR technology based on bibliometrics methods[J]. Curr. Biotechnol., 2022, 12(4): 606-613. |
| [1] | Yanyan JIA, Luyang DUANMU. Study on the Mechanism of Bubble Generation and Inhibition Method During Digital PCR Amplification Process [J]. Current Biotechnology, 2025, 15(4): 693-701. |
| [2] | Xin QI, Xinran LI, Yaning GUO, Dan WANG, Kai LI, Qiong WU, Liang LI. Comparison of Endogenous Genes in Maize Based on Digital PCR [J]. Current Biotechnology, 2025, 15(1): 78-85. |
| [3] | Hongbo FAN, Liangyong HU, Songqing HU. Establishment of Digital PCR Detection System for Helicobacter pyloriureC and 23S rDNA [J]. Current Biotechnology, 2024, 14(5): 868-874. |
| [4] | Kai LI, Jun FU, Rui CHEN, Xiaoyun CHEN, Liang LI. Transgenic Quantitative Detection Method Based on PCR-free High-throughput Sequencing Technology [J]. Current Biotechnology, 2024, 14(4): 610-617. |
| [5] | Enze CHENG-CHEN, Minghong JIA, Yueying LI. Research Progress of Nucleic Acid Reference Materials of Food-borne Pathogenic Bacteria [J]. Current Biotechnology, 2023, 13(2): 195-200. |
| [6] | Yan LI, Zhenzhou YANG, Wen LIANG. Development of Subsets CD4+ Proportion of T Lymphocyte Standard Reference Materials [J]. Current Biotechnology, 2022, 12(4): 600-605. |
| [7] | Peixuan MEN, Yufeng XIAO, Bin ZHANG. Analysis of Clinical Application and Technology Hotspots of Digital PCR Technology Based on Bibliometrics Methods [J]. Current Biotechnology, 2022, 12(4): 606-613. |
| [8] | LU Haiqiang1, JIAO Xinya1,2, WU Siyuan2, WANG Yali2, LIU Lu2, CHENG Shumei1, ZHANG Xiao3*, SU Xiaofeng2*. Application Progress on the Digital PCR in Detection of Foodborne Pathogenic Bacteria [J]. Curr. Biotech., 2021, 11(3): 260-268. |
| [9] | WANG Di, WU Xiao, WANG Zhidong, GAO Yunhua*. Research Progress on the Flow Cytometry Counting for Single Nucleotide Molecule [J]. Curr. Biotech., 2020, 10(6): 573-578. |
| [10] | ZHENG Zifan, LIU Fangfang, LIU Weixiao, JIN Wujun, LI Liang*. Application of Digital PCR Technology in the Development of Nucleic Acid Reference Materials [J]. Curr. Biotech., 2020, 10(6): 579-584. |
| [11] | WANG Shangjun1, DONG Lianhua2. Research on Calibration Method of Digital PCR Instrument [J]. Curr. Biotech., 2020, 10(6): 585-589. |
| [12] | YANG Jiayi1§*, CHEN Guifang2§, GAO Yunhuan1, WANG Zhidong1, WU Xiao1. Research Progress of Reference Material for HPV Nucleic Acid Detection [J]. Curr. Biotech., 2020, 10(6): 590-596. |
| [13] | ZHENG Zifan, LIU Weixiao, JIN Wujun, LI Liang*. Progress on Reference Materials Based on Mass Balance Method [J]. Curr. Biotech., 2020, 10(6): 623-629. |
| [14] | YAN Anxin1, CHEN Jing1, LIU Hongdi1, LI Yongjiu1, PENG Zhu1, ZHAO Hemiao1, LI Liang2, ZHAO Xingchun1*. Preparation of Plasmid DNA Reference Material Containing Human STR D6S1043-1 Fragment [J]. Curr. Biotech., 2020, 10(6): 630-636. |
| [15] | HU Sihong, YOU Guoye. Application Prospects of Digital PCR in Detection of SARS-CoV-2 [J]. Curr. Biotech., 2020, 10(6): 674-679. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||