Current Biotechnology ›› 2023, Vol. 13 ›› Issue (3): 412-424.DOI: 10.19586/j.2095-2341.2023.0007
• Articles • Previous Articles Next Articles
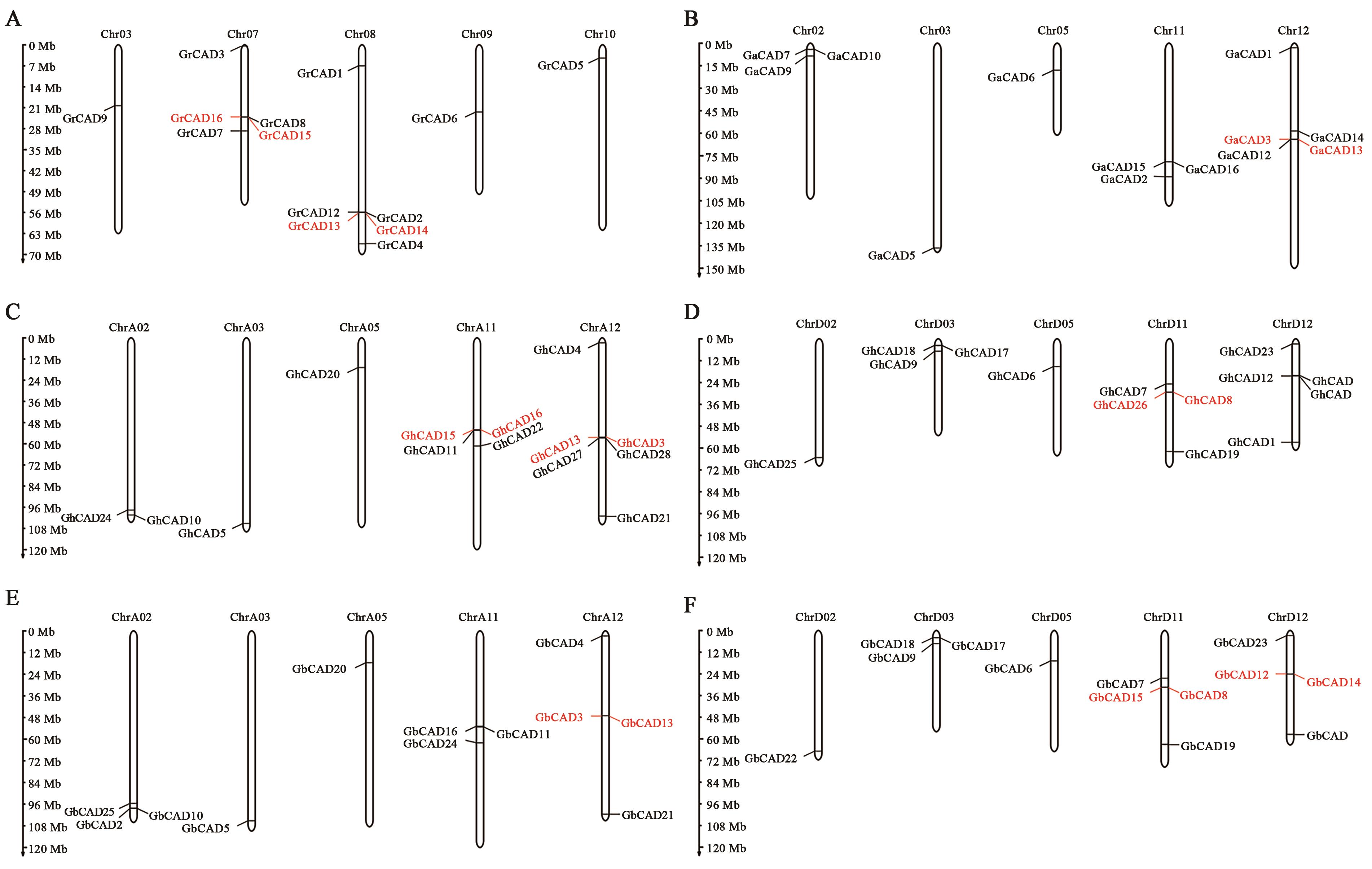
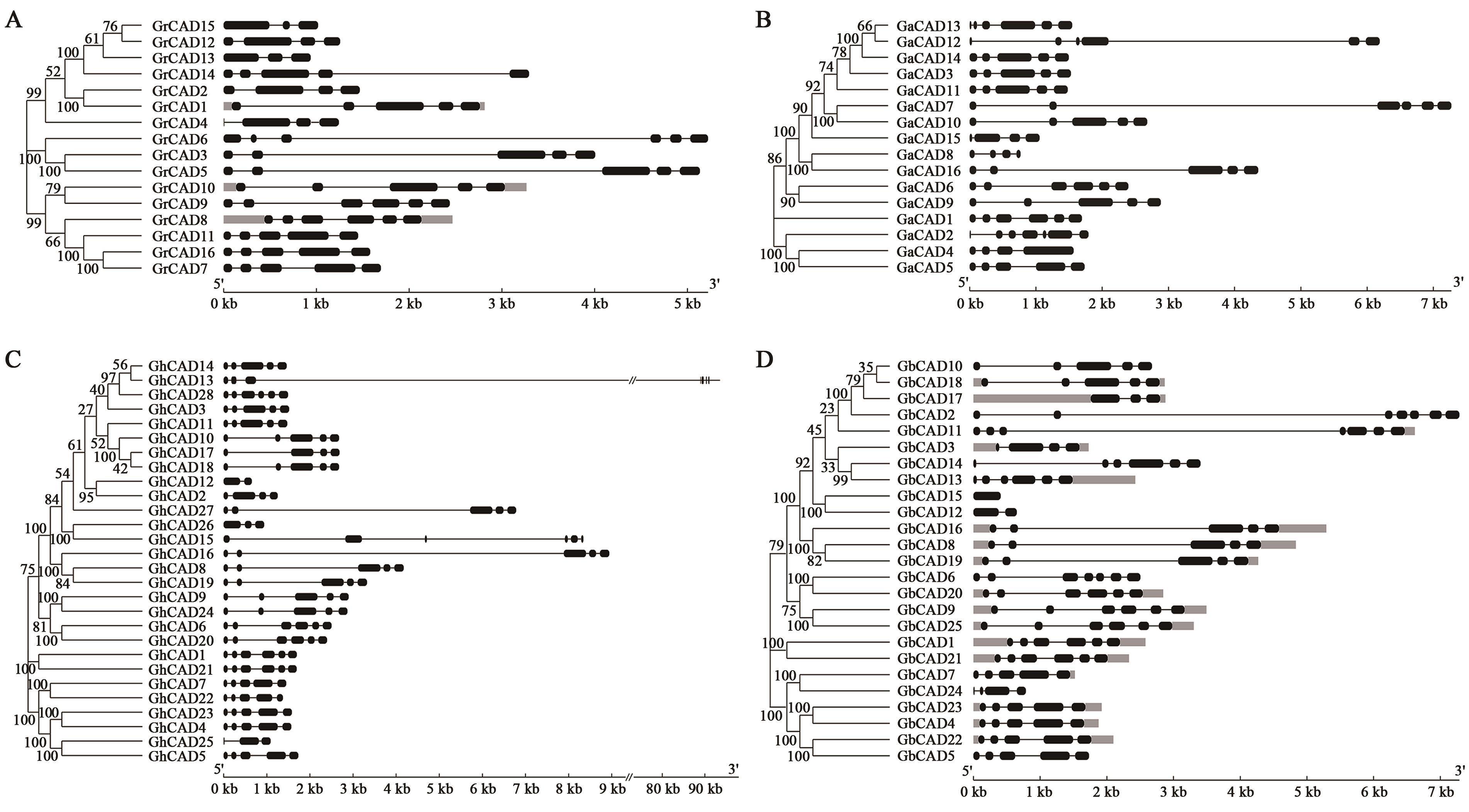
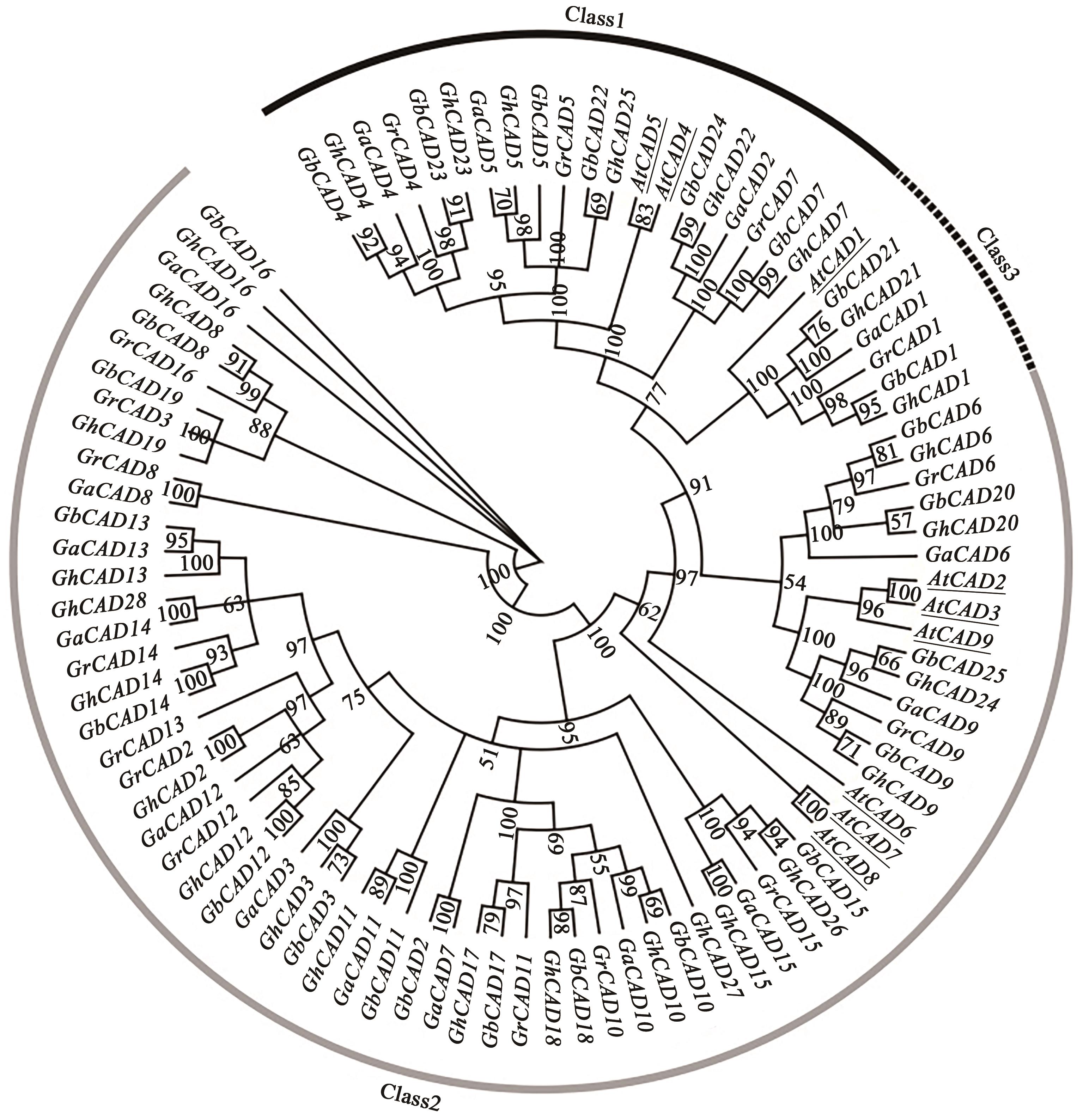
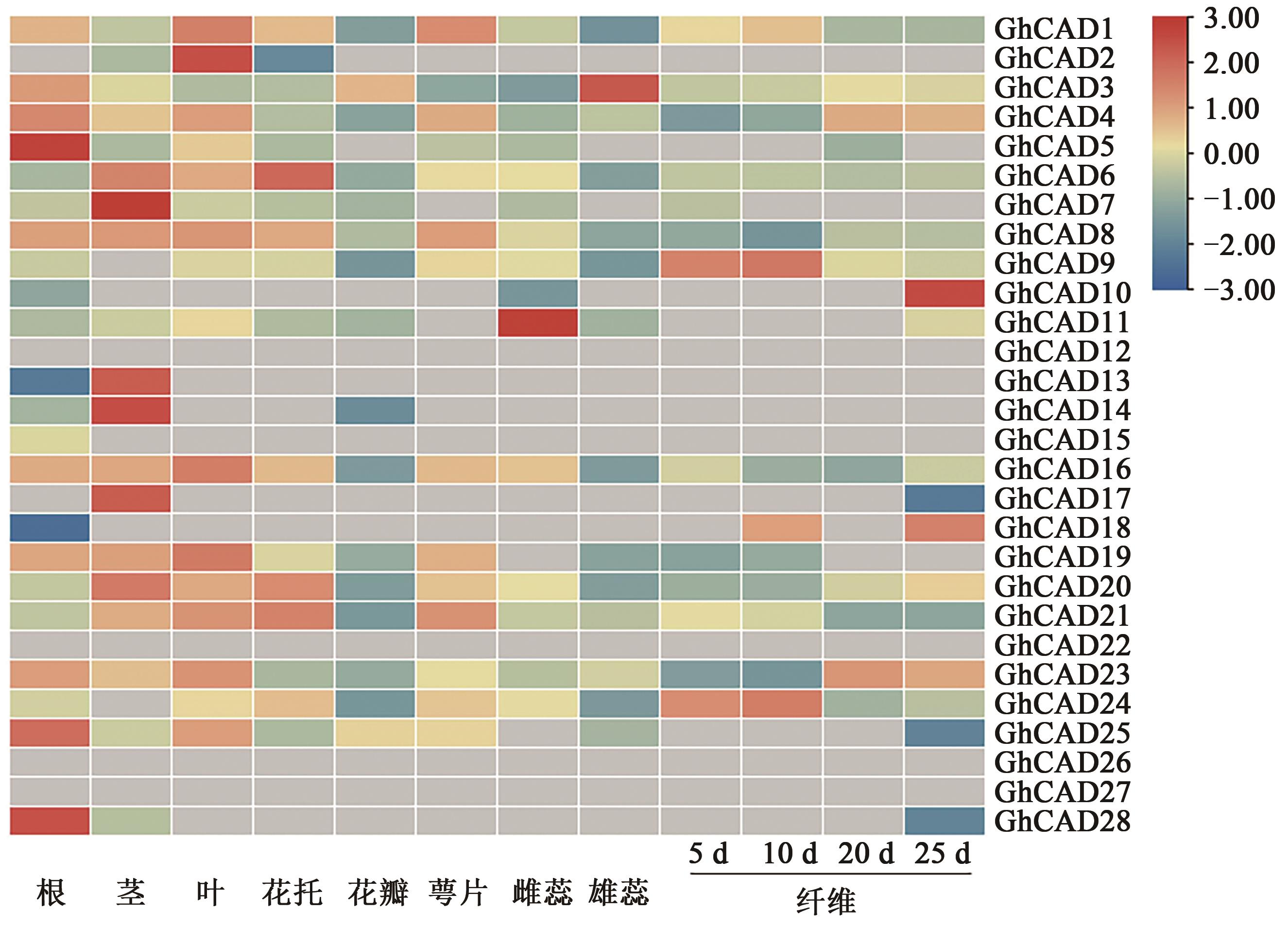
Bioinformatic Analysis of the CAD Gene Family in Cotton
Wenran HU1( ), Xiaoyan HAO1, Zhun ZHAO1, Wukui SHAO2, Shengqi GAO1, Jianping LI1, Quansheng HUANG1(
), Xiaoyan HAO1, Zhun ZHAO1, Wukui SHAO2, Shengqi GAO1, Jianping LI1, Quansheng HUANG1( )
)
- 1.Xinjiang Key Laboratory of Crop Biotechnology,Institute of Nuclear and Biological Technologies,Xinjiang Academy of Agricultural Sciences,Urumqi 830091,China
2.College of Life Sciences,Xinjiang Agricultural University,Urumqi 830052,China
-
Received:2023-02-02Accepted:2023-02-28Online:2023-05-25Published:2023-06-12 -
Contact:Quansheng HUANG
棉花CAD基因家族的生物信息学分析
胡文冉1( ), 郝晓燕1, 赵准1, 邵武奎2, 高升旗1, 李建平1, 黄全生1(
), 郝晓燕1, 赵准1, 邵武奎2, 高升旗1, 李建平1, 黄全生1( )
)
- 1.新疆农业科学院核技术生物技术研究所,新疆农作物生物技术重点实验室,乌鲁木齐 830091
2.新疆农业大学生命科学学院,乌鲁木齐 830052
-
通讯作者:黄全生 -
作者简介:胡文冉 E-mail: huwran@126.com; -
基金资助:新疆维吾尔自治区自然科学基金资助项目(2022D01A88);新疆维吾尔自治区重点实验室开放课题(2021D04002);新疆农作物生物技术重点实验室开放课题(XJYS0302-2020-02)
CLC Number:
Cite this article
Wenran HU, Xiaoyan HAO, Zhun ZHAO, Wukui SHAO, Shengqi GAO, Jianping LI, Quansheng HUANG. Bioinformatic Analysis of the CAD Gene Family in Cotton[J]. Current Biotechnology, 2023, 13(3): 412-424.
胡文冉, 郝晓燕, 赵准, 邵武奎, 高升旗, 李建平, 黄全生. 棉花CAD基因家族的生物信息学分析[J]. 生物技术进展, 2023, 13(3): 412-424.
share this article
| 基因 | 基因组登陆号 | 染色体位置 | 编码序列长度/ bp | 蛋白长度/ aa | 分子量/ kD | 等电点 pI | 亚细胞定位 |
|---|---|---|---|---|---|---|---|
| GrCAD1 | Cotton_D_gene_10016775 | Chr08 | 1 068 | 355 | 38.45 | 6.65 | 细胞质 |
| GrCAD2 | Cotton_D_gene_10037629 | Chr08 | 942 | 313 | 33.98 | 6.31 | 细胞质 |
| GrCAD3 | Cotton_D_gene_10012650 | Chr07 | 1 095 | 364 | 39.12 | 5.85 | 细胞质 |
| GrCAD4 | Cotton_D_gene_10038273 | Chr08 | 1 074 | 357 | 38.71 | 5.41 | 细胞质 |
| GrCAD5 | Cotton_D_gene_10016445 | Chr10 | 1 077 | 358 | 38.70 | 5.64 | 细胞质 |
| GrCAD6 | Cotton_D_gene_10033972 | Chr09 | 1 080 | 359 | 39.19 | 7.13 | 细胞质 |
| GrCAD7 | Cotton_D_gene_10036902 | Chr07 | 1 059 | 352 | 38.18 | 6.12 | 细胞质 |
| GrCAD8 | Cotton_D_gene_10014351 | Chr07 | 780 | 259 | 28.15 | 5.99 | 细胞质 |
| GrCAD9 | Cotton_D_gene_10036599 | Chr03 | 1 080 | 359 | 39.09 | 5.58 | 细胞质 |
| GrCAD10 | Cotton_D_gene_10002391 | scaffold369 | 990 | 329 | 35.61 | 6.07 | 细胞质 |
| GrCAD11 | Cotton_D_gene_10002389 | scaffold369 | 1 086 | 361 | 39.29 | 6.26 | 细胞质 |
| GrCAD12 | Cotton_D_gene_10037630 | Chr08 | 741 | 246 | 27.09 | 8.94 | 细胞质 |
| GrCAD13 | Cotton_D_gene_10037632 | Chr08 | 774 | 257 | 28.02 | 8.12 | 细胞质 |
| GrCAD14 | Cotton_D_gene_10037631 | Chr08 | 1 089 | 362 | 39.10 | 6.50 | 细胞质 |
| GrCAD15 | Cotton_D_gene_10014346 | Chr07 | 882 | 293 | 31.46 | 6.25 | 细胞质 |
| GrCAD16 | Cotton_D_gene_10014349 | Chr07 | 1 095 | 364 | 39.11 | 5.75 | 细胞质 |
Table 1 The characteristic of CAD genes in Gossypium raimondii
| 基因 | 基因组登陆号 | 染色体位置 | 编码序列长度/ bp | 蛋白长度/ aa | 分子量/ kD | 等电点 pI | 亚细胞定位 |
|---|---|---|---|---|---|---|---|
| GrCAD1 | Cotton_D_gene_10016775 | Chr08 | 1 068 | 355 | 38.45 | 6.65 | 细胞质 |
| GrCAD2 | Cotton_D_gene_10037629 | Chr08 | 942 | 313 | 33.98 | 6.31 | 细胞质 |
| GrCAD3 | Cotton_D_gene_10012650 | Chr07 | 1 095 | 364 | 39.12 | 5.85 | 细胞质 |
| GrCAD4 | Cotton_D_gene_10038273 | Chr08 | 1 074 | 357 | 38.71 | 5.41 | 细胞质 |
| GrCAD5 | Cotton_D_gene_10016445 | Chr10 | 1 077 | 358 | 38.70 | 5.64 | 细胞质 |
| GrCAD6 | Cotton_D_gene_10033972 | Chr09 | 1 080 | 359 | 39.19 | 7.13 | 细胞质 |
| GrCAD7 | Cotton_D_gene_10036902 | Chr07 | 1 059 | 352 | 38.18 | 6.12 | 细胞质 |
| GrCAD8 | Cotton_D_gene_10014351 | Chr07 | 780 | 259 | 28.15 | 5.99 | 细胞质 |
| GrCAD9 | Cotton_D_gene_10036599 | Chr03 | 1 080 | 359 | 39.09 | 5.58 | 细胞质 |
| GrCAD10 | Cotton_D_gene_10002391 | scaffold369 | 990 | 329 | 35.61 | 6.07 | 细胞质 |
| GrCAD11 | Cotton_D_gene_10002389 | scaffold369 | 1 086 | 361 | 39.29 | 6.26 | 细胞质 |
| GrCAD12 | Cotton_D_gene_10037630 | Chr08 | 741 | 246 | 27.09 | 8.94 | 细胞质 |
| GrCAD13 | Cotton_D_gene_10037632 | Chr08 | 774 | 257 | 28.02 | 8.12 | 细胞质 |
| GrCAD14 | Cotton_D_gene_10037631 | Chr08 | 1 089 | 362 | 39.10 | 6.50 | 细胞质 |
| GrCAD15 | Cotton_D_gene_10014346 | Chr07 | 882 | 293 | 31.46 | 6.25 | 细胞质 |
| GrCAD16 | Cotton_D_gene_10014349 | Chr07 | 1 095 | 364 | 39.11 | 5.75 | 细胞质 |
| 基因 | 基因组登陆号 | 染色体位置 | 编码序列长度/ bp | 蛋白长度/ aa | 分子量/ kD | 等电点 pI | 亚细胞定位 |
|---|---|---|---|---|---|---|---|
| GaCAD1 | Ga12G0361 | Chr12 | 1 068 | 355 | 38.41 | 7.05 | 细胞质 |
| GaCAD2 | Ga11G1886 | Chr11 | 987 | 328 | 35.17 | 6.56 | 细胞质 |
| GaCAD3 | Ga12G2139 | Chr12 | 1 098 | 365 | 39.70 | 6.22 | 细胞质 |
| GaCAD4 | Ga14G0143 | tig00000498 | 1 188 | 395 | 42.93 | 5.26 | 细胞质 |
| GaCAD5 | Ga03G2431 | Chr03 | 1 077 | 358 | 38.84 | 5.53 | 细胞质 |
| GaCAD6 | Ga05G1953 | Chr05 | 1 074 | 357 | 39.12 | 6.46 | 细胞质 |
| GaCAD7 | Ga02G0337 | Chr02 | 1 065 | 354 | 39.74 | 6.13 | 细胞质 |
| GaCAD8 | Ga14G1938 | tig00016738 | 339 | 112 | 11.76 | 9.10 | 细胞质 |
| GaCAD9 | Ga02G0499 | Chr02 | 1 080 | 359 | 39.07 | 6.01 | 细胞质 |
| GaCAD10 | Ga02G0339 | Chr02 | 1 086 | 361 | 39.22 | 6.26 | 细胞质 |
| GaCAD11 | Ga14G1937 | tig00016738 | 1 086 | 361 | 39.07 | 7.61 | 细胞质 |
| GaCAD12 | Ga12G2141 | Chr12 | 945 | 314 | 34.10 | 6.45 | 细胞质 |
| GaCAD13 | Ga12G2140 | Chr12 | 1 065 | 354 | 37.94 | 7.15 | 细胞质 |
| GaCAD14 | Ga12G2105 | Chr12 | 1 107 | 368 | 39.62 | 6.64 | 细胞质 |
| GaCAD15 | Ga11G1713 | Chr11 | 783 | 260 | 28.16 | 7.17 | 细胞质 |
| GaCAD16 | Ga11G1714 | Chr11 | 1 095 | 364 | 39.13 | 5.75 | 细胞质 |
Table 2 The characteristic of CAD genes in Gossypium arboretum
| 基因 | 基因组登陆号 | 染色体位置 | 编码序列长度/ bp | 蛋白长度/ aa | 分子量/ kD | 等电点 pI | 亚细胞定位 |
|---|---|---|---|---|---|---|---|
| GaCAD1 | Ga12G0361 | Chr12 | 1 068 | 355 | 38.41 | 7.05 | 细胞质 |
| GaCAD2 | Ga11G1886 | Chr11 | 987 | 328 | 35.17 | 6.56 | 细胞质 |
| GaCAD3 | Ga12G2139 | Chr12 | 1 098 | 365 | 39.70 | 6.22 | 细胞质 |
| GaCAD4 | Ga14G0143 | tig00000498 | 1 188 | 395 | 42.93 | 5.26 | 细胞质 |
| GaCAD5 | Ga03G2431 | Chr03 | 1 077 | 358 | 38.84 | 5.53 | 细胞质 |
| GaCAD6 | Ga05G1953 | Chr05 | 1 074 | 357 | 39.12 | 6.46 | 细胞质 |
| GaCAD7 | Ga02G0337 | Chr02 | 1 065 | 354 | 39.74 | 6.13 | 细胞质 |
| GaCAD8 | Ga14G1938 | tig00016738 | 339 | 112 | 11.76 | 9.10 | 细胞质 |
| GaCAD9 | Ga02G0499 | Chr02 | 1 080 | 359 | 39.07 | 6.01 | 细胞质 |
| GaCAD10 | Ga02G0339 | Chr02 | 1 086 | 361 | 39.22 | 6.26 | 细胞质 |
| GaCAD11 | Ga14G1937 | tig00016738 | 1 086 | 361 | 39.07 | 7.61 | 细胞质 |
| GaCAD12 | Ga12G2141 | Chr12 | 945 | 314 | 34.10 | 6.45 | 细胞质 |
| GaCAD13 | Ga12G2140 | Chr12 | 1 065 | 354 | 37.94 | 7.15 | 细胞质 |
| GaCAD14 | Ga12G2105 | Chr12 | 1 107 | 368 | 39.62 | 6.64 | 细胞质 |
| GaCAD15 | Ga11G1713 | Chr11 | 783 | 260 | 28.16 | 7.17 | 细胞质 |
| GaCAD16 | Ga11G1714 | Chr11 | 1 095 | 364 | 39.13 | 5.75 | 细胞质 |
| 基因 | 基因组登陆号 | 染色体位置 | 编码序列长度/ bp | 蛋白长度/ aa | 分子量/ kD | 等电点 pI | 亚细胞定位 |
|---|---|---|---|---|---|---|---|
| GhCAD1 | GH_D12G2651 | Chr12 | 1 068 | 355 | 38.45 | 7.52 | 细胞质 |
| GhCAD2 | GH_D12G0806 | Chr12 | 945 | 314 | 33.66 | 6.59 | 细胞质 |
| GhCAD3 | GH_A12G0990 | Chr12 | 1 086 | 361 | 39.24 | 6.60 | 细胞质 |
| GhCAD4 | GH_A12G0204 | Chr12 | 1 074 | 357 | 38.76 | 5.32 | 细胞质 |
| GhCAD5 | GH_A03G2100 | Chr03 | 1 077 | 358 | 38.81 | 5.53 | 细胞质 |
| GhCAD6 | GH_D05G1877 | Chr05 | 1 080 | 359 | 39.18 | 7.13 | 细胞质 |
| GhCAD7 | GH_D11G2110 | Chr11 | 1 059 | 352 | 38.19 | 6.01 | 细胞质 |
| GhCAD8 | GH_D11G2243 | Chr11 | 1 095 | 364 | 39.14 | 5.85 | 细胞质 |
| GhCAD9 | GH_D03G0499 | Chr03 | 1 080 | 359 | 39.09 | 5.85 | 细胞质 |
| GhCAD10 | GH_A02G1713 | Chr02 | 1 086 | 361 | 39.28 | 6.00 | 细胞质 |
| GhCAD11 | GH_A11G2195 | Chr11 | 1 086 | 361 | 39.07 | 7.61 | 细胞质 |
| GhCAD12 | GH_D12G0807 | Chr12 | 549 | 182 | 19.91 | 7.65 | 细胞质 |
| GhCAD13 | GH_A12G0991 | Chr12 | 1 398 | 465 | 50.54 | 6.02 | 细胞质 |
| GhCAD14 | GH_D12G0808 | Chr12 | 1 089 | 362 | 38.99 | 6.46 | 细胞质 |
| GhCAD15 | GH_A11G2192 | Chr11 | 831 | 276 | 29.86 | 6.59 | 细胞质 |
| GhCAD16 | GH_A11G2193 | Chr11 | 1 095 | 364 | 39.13 | 5.75 | 细胞质 |
| GhCAD17 | GH_D03G0349 | Chr03 | 972 | 323 | 34.96 | 6.71 | 细胞质 |
| GhCAD18 | GH_D03G0347 | Chr03 | 1 086 | 361 | 39.19 | 6.12 | 细胞质 |
| GhCAD19 | GH_D11G3241 | Chr11 | 1 095 | 364 | 39.13 | 5.85 | 细胞质 |
| GhCAD20 | GH_A05G1840 | Chr05 | 1 080 | 359 | 39.05 | 6.41 | 细胞质 |
| GhCAD21 | GH_A12G2629 | Chr12 | 1 068 | 355 | 38.38 | 7.05 | 细胞质 |
| GhCAD22 | GH_A11G2344 | Chr11 | 894 | 297 | 31.97 | 6.23 | 细胞质 |
| GhCAD23 | GH_D12G0217 | Chr12 | 1 074 | 357 | 38.71 | 5.41 | 细胞质 |
| GhCAD24 | GH_A02G1628 | Chr02 | 1 080 | 359 | 39.07 | 5.71 | 细胞质 |
| GhCAD25 | GH_D02G2271 | Chr02 | 663 | 220 | 24.01 | 5.70 | 细胞质 |
| GhCAD26 | GH_D11G2246 | Chr11 | 747 | 248 | 26.73 | 7.14 | 细胞质 |
| GhCAD27 | GH_A12G0994 | Chr12 | 1 137 | 378 | 40.94 | 6.49 | 细胞质 |
| GhCAD28 | GH_A12G0993 | Chr12 | 1 014 | 337 | 36.49 | 6.08 | 细胞质 |
Table 3 The characteristic of CAD genes in Gossypium hirsutum
| 基因 | 基因组登陆号 | 染色体位置 | 编码序列长度/ bp | 蛋白长度/ aa | 分子量/ kD | 等电点 pI | 亚细胞定位 |
|---|---|---|---|---|---|---|---|
| GhCAD1 | GH_D12G2651 | Chr12 | 1 068 | 355 | 38.45 | 7.52 | 细胞质 |
| GhCAD2 | GH_D12G0806 | Chr12 | 945 | 314 | 33.66 | 6.59 | 细胞质 |
| GhCAD3 | GH_A12G0990 | Chr12 | 1 086 | 361 | 39.24 | 6.60 | 细胞质 |
| GhCAD4 | GH_A12G0204 | Chr12 | 1 074 | 357 | 38.76 | 5.32 | 细胞质 |
| GhCAD5 | GH_A03G2100 | Chr03 | 1 077 | 358 | 38.81 | 5.53 | 细胞质 |
| GhCAD6 | GH_D05G1877 | Chr05 | 1 080 | 359 | 39.18 | 7.13 | 细胞质 |
| GhCAD7 | GH_D11G2110 | Chr11 | 1 059 | 352 | 38.19 | 6.01 | 细胞质 |
| GhCAD8 | GH_D11G2243 | Chr11 | 1 095 | 364 | 39.14 | 5.85 | 细胞质 |
| GhCAD9 | GH_D03G0499 | Chr03 | 1 080 | 359 | 39.09 | 5.85 | 细胞质 |
| GhCAD10 | GH_A02G1713 | Chr02 | 1 086 | 361 | 39.28 | 6.00 | 细胞质 |
| GhCAD11 | GH_A11G2195 | Chr11 | 1 086 | 361 | 39.07 | 7.61 | 细胞质 |
| GhCAD12 | GH_D12G0807 | Chr12 | 549 | 182 | 19.91 | 7.65 | 细胞质 |
| GhCAD13 | GH_A12G0991 | Chr12 | 1 398 | 465 | 50.54 | 6.02 | 细胞质 |
| GhCAD14 | GH_D12G0808 | Chr12 | 1 089 | 362 | 38.99 | 6.46 | 细胞质 |
| GhCAD15 | GH_A11G2192 | Chr11 | 831 | 276 | 29.86 | 6.59 | 细胞质 |
| GhCAD16 | GH_A11G2193 | Chr11 | 1 095 | 364 | 39.13 | 5.75 | 细胞质 |
| GhCAD17 | GH_D03G0349 | Chr03 | 972 | 323 | 34.96 | 6.71 | 细胞质 |
| GhCAD18 | GH_D03G0347 | Chr03 | 1 086 | 361 | 39.19 | 6.12 | 细胞质 |
| GhCAD19 | GH_D11G3241 | Chr11 | 1 095 | 364 | 39.13 | 5.85 | 细胞质 |
| GhCAD20 | GH_A05G1840 | Chr05 | 1 080 | 359 | 39.05 | 6.41 | 细胞质 |
| GhCAD21 | GH_A12G2629 | Chr12 | 1 068 | 355 | 38.38 | 7.05 | 细胞质 |
| GhCAD22 | GH_A11G2344 | Chr11 | 894 | 297 | 31.97 | 6.23 | 细胞质 |
| GhCAD23 | GH_D12G0217 | Chr12 | 1 074 | 357 | 38.71 | 5.41 | 细胞质 |
| GhCAD24 | GH_A02G1628 | Chr02 | 1 080 | 359 | 39.07 | 5.71 | 细胞质 |
| GhCAD25 | GH_D02G2271 | Chr02 | 663 | 220 | 24.01 | 5.70 | 细胞质 |
| GhCAD26 | GH_D11G2246 | Chr11 | 747 | 248 | 26.73 | 7.14 | 细胞质 |
| GhCAD27 | GH_A12G0994 | Chr12 | 1 137 | 378 | 40.94 | 6.49 | 细胞质 |
| GhCAD28 | GH_A12G0993 | Chr12 | 1 014 | 337 | 36.49 | 6.08 | 细胞质 |
| 基因 | 基因组登陆号 | 染色体位置 | 编码序列长度/ bp | 蛋白长度/ aa | 分子量/ kD | 等电点 pI | 亚细胞定位 |
|---|---|---|---|---|---|---|---|
| GbCAD1 | Gbar_D12G025360 | Chr12 | 1 068 | 355 | 38.45 | 7.52 | 细胞质 |
| GbCAD2 | Gbar_A02G015780 | Chr02 | 981 | 326 | 36.30 | 6.21 | 细胞质 |
| GbCAD3 | Gbar_A12G009020 | Chr12 | 921 | 306 | 33.01 | 6.25 | 细胞质 |
| GbCAD4 | Gbar_A12G002060 | Chr12 | 1 074 | 357 | 38.76 | 5.32 | 细胞质 |
| GbCAD5 | Gbar_A03G020480 | Chr03 | 1 077 | 358 | 38.82 | 5.53 | 细胞质 |
| GbCAD6 | Gbar_D05G018810 | Chr05 | 1 035 | 344 | 37.76 | 6.55 | 细胞质 |
| GbCAD7 | Gbar_D11G021150 | Chr11 | 1 059 | 352 | 38.19 | 6.01 | 细胞质 |
| GbCAD8 | Gbar_D11G022440 | Chr11 | 1 095 | 364 | 39.14 | 5.85 | 细胞质 |
| GbCAD9 | Gbar_D03G004820 | Chr03 | 984 | 327 | 35.62 | 6.19 | 细胞质 |
| GbCAD10 | Gbar_A02G015760 | Chr02 | 1 086 | 361 | 39.33 | 6.26 | 细胞质 |
| GbCAD11 | Gbar_A11G021420 | Chr11 | 1 056 | 351 | 37.98 | 8.63 | 细胞质 |
| GbCAD12 | Gbar_D12G008930 | Chr12 | 549 | 182 | 19.94 | 7.65 | 细胞质 |
| GbCAD13 | Gbar_A12G009010 | Chr12 | 978 | 325 | 34.74 | 6.64 | 细胞质 |
| GbCAD14 | Gbar_D12G008920 | Chr12 | 1 122 | 373 | 40.12 | 6.23 | 细胞质 |
| GbCAD15 | Gbar_D11G022460 | Chr11 | 408 | 135 | 14.46 | 5.62 | 细胞质 |
| GbCAD16 | Gbar_A11G021400 | Chr11 | 1 095 | 364 | 39.13 | 5.75 | 细胞质 |
| GbCAD17 | Gbar_D03G003450 | Chr03 | 792 | 263 | 28.34 | 6.15 | 细胞质 |
| GbCAD18 | Gbar_D03G003430 | Chr03 | 1 086 | 361 | 39.16 | 6.12 | 细胞质 |
| GbCAD19 | Gbar_D11G031180 | Chr11 | 1 098 | 365 | 39.26 | 5.75 | 细胞质 |
| GbCAD20 | Gbar_A05G018170 | Chr05 | 1 080 | 359 | 39.17 | 6.82 | 细胞质 |
| GbCAD21 | Gbar_A12G025440 | Chr12 | 1 068 | 355 | 38.38 | 7.05 | 细胞质 |
| GbCAD22 | Gbar_D02G022410 | Chr02 | 1 077 | 358 | 38.82 | 5.64 | 细胞质 |
| GbCAD23 | Gbar_D12G002300 | Chr12 | 1 074 | 357 | 38.71 | 5.41 | 细胞质 |
| GbCAD24 | Gbar_A11G022760 | Chr11 | 561 | 186 | 19.77 | 7.99 | 细胞质 |
| GbCAD25 | Gbar_A02G014990 | Chr02 | 996 | 331 | 36.10 | 6.46 | 细胞质 |
Table 4 The characteristic of CAD genes in Gossypium barbadense
| 基因 | 基因组登陆号 | 染色体位置 | 编码序列长度/ bp | 蛋白长度/ aa | 分子量/ kD | 等电点 pI | 亚细胞定位 |
|---|---|---|---|---|---|---|---|
| GbCAD1 | Gbar_D12G025360 | Chr12 | 1 068 | 355 | 38.45 | 7.52 | 细胞质 |
| GbCAD2 | Gbar_A02G015780 | Chr02 | 981 | 326 | 36.30 | 6.21 | 细胞质 |
| GbCAD3 | Gbar_A12G009020 | Chr12 | 921 | 306 | 33.01 | 6.25 | 细胞质 |
| GbCAD4 | Gbar_A12G002060 | Chr12 | 1 074 | 357 | 38.76 | 5.32 | 细胞质 |
| GbCAD5 | Gbar_A03G020480 | Chr03 | 1 077 | 358 | 38.82 | 5.53 | 细胞质 |
| GbCAD6 | Gbar_D05G018810 | Chr05 | 1 035 | 344 | 37.76 | 6.55 | 细胞质 |
| GbCAD7 | Gbar_D11G021150 | Chr11 | 1 059 | 352 | 38.19 | 6.01 | 细胞质 |
| GbCAD8 | Gbar_D11G022440 | Chr11 | 1 095 | 364 | 39.14 | 5.85 | 细胞质 |
| GbCAD9 | Gbar_D03G004820 | Chr03 | 984 | 327 | 35.62 | 6.19 | 细胞质 |
| GbCAD10 | Gbar_A02G015760 | Chr02 | 1 086 | 361 | 39.33 | 6.26 | 细胞质 |
| GbCAD11 | Gbar_A11G021420 | Chr11 | 1 056 | 351 | 37.98 | 8.63 | 细胞质 |
| GbCAD12 | Gbar_D12G008930 | Chr12 | 549 | 182 | 19.94 | 7.65 | 细胞质 |
| GbCAD13 | Gbar_A12G009010 | Chr12 | 978 | 325 | 34.74 | 6.64 | 细胞质 |
| GbCAD14 | Gbar_D12G008920 | Chr12 | 1 122 | 373 | 40.12 | 6.23 | 细胞质 |
| GbCAD15 | Gbar_D11G022460 | Chr11 | 408 | 135 | 14.46 | 5.62 | 细胞质 |
| GbCAD16 | Gbar_A11G021400 | Chr11 | 1 095 | 364 | 39.13 | 5.75 | 细胞质 |
| GbCAD17 | Gbar_D03G003450 | Chr03 | 792 | 263 | 28.34 | 6.15 | 细胞质 |
| GbCAD18 | Gbar_D03G003430 | Chr03 | 1 086 | 361 | 39.16 | 6.12 | 细胞质 |
| GbCAD19 | Gbar_D11G031180 | Chr11 | 1 098 | 365 | 39.26 | 5.75 | 细胞质 |
| GbCAD20 | Gbar_A05G018170 | Chr05 | 1 080 | 359 | 39.17 | 6.82 | 细胞质 |
| GbCAD21 | Gbar_A12G025440 | Chr12 | 1 068 | 355 | 38.38 | 7.05 | 细胞质 |
| GbCAD22 | Gbar_D02G022410 | Chr02 | 1 077 | 358 | 38.82 | 5.64 | 细胞质 |
| GbCAD23 | Gbar_D12G002300 | Chr12 | 1 074 | 357 | 38.71 | 5.41 | 细胞质 |
| GbCAD24 | Gbar_A11G022760 | Chr11 | 561 | 186 | 19.77 | 7.99 | 细胞质 |
| GbCAD25 | Gbar_A02G014990 | Chr02 | 996 | 331 | 36.10 | 6.46 | 细胞质 |
| 1 | BOERJAN W, RALPH J. BAUCHER M. Lignin biosynthesis[J]. Annu. Rev. Plant Biol., 2003, 54: 519-546. |
| 2 | GOU J Y, WANG L J, CHEN S P, et al.. Gene expression and metabolite profiles of cotton fiber during cell elongation and secondary cell wall synthesis[J]. Cell Res., 2007, 17: 422-434. |
| 3 | 尚军,吴旺泽,马永贵.植物苯丙烷代谢途径[J].中国生物化学与分子生物学报,2022,38(11):1467-1476. |
| 4 | FAN L, SHI W J, HU W R, et al.. Molecular and biochemical evidence for phenylpropanoid synthesis and presence of wall-linked phenolics in cotton fibers[J]. J. Integr. Plant Biol., 2009, 51(7): 626-637. |
| 5 | KIM S J, KIM M R, BEDGAR D L, et al.. Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis [J]. Prco. Natl. Acad. Sci. USA, 2004, 101(6): 1455-1460. |
| 6 | TOBIAS C M, CHOW E K. Structure of the cinnamyl-alcohol dehydrogenase gene family in rice and promoter activity of a member associated with lignification[J]. Planta, 2005, 220(5): 678-688. |
| 7 | SABALLOS A, EJETA G, SANCHEZ E, et al.. A genomewide analysis of the cinnamyl alcohol dehydrogenase family in sorghum [Sorghum bicolor (L.) Moench] identifies SbCAD2 as the brown midrib6 gene[J]. Genetics, 2009, 181(2): 783-795. |
| 8 | KNIGHT M E, HALPIN C, SCHUCH W. Identification and characterization of cDNA clones encoding cinnamyl alcohol dehydrogenase from tobacco[J]. Plant Mol. Biol., 1992, 19(5): 793-801. |
| 9 | SIBOUT R, EUDES A, MOUILLE G, et al.. cinnamyl alcohol dehydrogenase-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis [J]. Plant Cell, 2005, 17(7): 2059-2076. |
| 10 | KIM S J, KIM K W, CHO M H, et al.. Expression of cinnamyl alcohol dehydrogenase and their putative homologues during Arabidopsis thaliana growth and development: lessons for database annotations?[J]. Phytochemistry, 2011, 68(14): 1957-1974. |
| 11 | LI L G, CHENG X F, LESHKEVICH J, et al.. The last step of syringyl monolignol biosynthesis in angiosperms is regulated by a novel gene encoding sinapyl alcohol dehydrogenase[J]. Plant Cell, 2001, 13(7): 1567-1586. |
| 12 | GUILLAUMIE S, PICHON M, MARTINANT J P, et al. Differential expression of phenylpropanoid and related genes in brown-midrib bm1, bm2, bm3 and bm4 young near-isogenic maize plants[J]. Planta, 2007, 226: 235-250. |
| 13 | ZHANG K, QIAN Q, HUANG Z, et al.. GOLD HULL AND INTERNODE2 encodes a primary multifunction cinnamyl-alcohol dehydrogenase in rice[J]. Plant Physiol., 2006, 140(3): 972-983. |
| 14 | GUO D M, RAN J H, WANG X Q. Evolution of the cinnamyl/sinapyl alcohol dehydrogenase (CAD/SAD) gene family: the emergence of real lignin is associated with the origin of bona fide CAD [J]. J. Mol. Evol., 2010, 71: 202-218. |
| 15 | 张鲁斌,谷会,弓德强,等.植物肉桂醇脱氢酶及其基因研究进展[J].西北植物学报,2011,31(1):204-211. |
| 16 | SCHUBERT R, SPERISEN C, MÜLLER-STARCK G, et al.. The cinnamyl alcohol dehydrogenase gene structure in Picea abies (L.)Karst: genomic sequences, southern hybridization, genetic analysis and phylogenetic relationships[J]. Trees, 1998, 12: 453-463. |
| 17 | QI K J, SONG X F, YUAN Y Z, et al.. CAD genes: genome-wide identification, evolution, and their contribution to lignin biosynthesis in pear (Pyrus bretschneideri)[J/OL]. Plants, 2021, 10: 1444[2021-07-15]. . |
| 18 | EL-GEBALI S, MISTRY J, BATEMAN A, et al.. The pfam protein families database in 2019[J]. Nucl. Acids Res., 2019, 47(D1): 427-432. |
| 19 | EOM H S, KIM H, HYUN K T. The cinnamyl alcohol dehydrogenase (CAD) gene family in flax (Linum usitatissimum L.): insight from expression profiling of cads induced by elicitors in cultured flax cells[J]. Arch. Biol. Sci., 2016, 68(3): 603-612. |
| 20 | 高正银,孙文杰,宋晓云,等.雷蒙德棉第Ⅲ类过氧化物酶全基因组鉴定和表达分析[J].生物技术进展,2019,9(5):490-501. |
| 21 | ZHANG T Z, HU Y, JIANG W K, et al.. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement[J]. Nat. Biotechnol., 2015, 33: 531-537. |
| 22 | CANNON S B, MITRA A, BAUMGARTEN A, et al.. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana [J/OL]. BMC Plant Biol., 2004, 4: 10[2004-06-01]. . |
| 23 | HISANO H, NANDAKUMAR R, WANG Z Y. Genetic modification of lignin biosynthesis for improved biofuel production[J]. In Vitro Cell Dev. Biol. Plant, 2009, 45: 306-313. |
| 24 | HOVAV R, UDALL J A, HOVAV E, et al.. A majority of cotton genes are expressed in single-celled fiber[J]. Planta, 2008, 227: 319-329. |
| 25 | HU W R, LINKER R, FAN L, et al.. Accumulation and structural unit of wall-linked phenolics in the cotton (Gossypium hirsutum) fiber secondary wall[J]. Int. J. Agric. Biol., 2019, 21: 1063-1072. |
| 26 | TRONCHET M, BALAGUÉ C, KROJ T, et al.. Cinnamyl alcohol dehydrogenases-C and D, key enzymes in lignin biosynthesis, play an essential role in disease resistance in Arabidopsis [J]. Mol. Plant Pathol., 2010, 11(1): 83-92. |
| 27 | SIBOUT R, EUDES A, POLLET B, et al.. Expression pattern of two paralogs encoding cinnamyl alcohol dehydrogenases in Arabidopsis isolation and characterization of the corresponding mutants[J]. Plant Physiol., 2003, 132: 848-860. |
| 28 | RAES J, ROHDE A, CHRISTENSEN J H, et al.. Genome-wide characterization of the lignification toolbox in Arabidopsis [J]. Plant Physiol., 2003, 133(3): 1051-1071. |
| 29 | LI X M, YUAN D J, ZHANG J F, et al.. Genetic mapping and characteristics of genes specifically or preferentially expressed during fiber development in cotton[J/OL]. PLoS ONE, 2013, 8: e54444[2013-01-25]. . |
| 30 | 胡文冉,李晓东,周小云,等.棉花GhCAD6基因表达及其功能分析研究[J].生物技术进展,2019, 9(1):46-53. |
| 31 | CAROCHA V, SOLER M, HEFER C, et al.. Genome-wide analysis of the lignin toolbox of Eucalyptus grandis [J]. New Phytol., 2015, 206: 1297-1313. |
| 32 | HIRANO K, AYA K, KONDO M, et al.. OsCAD2 is the major CAD gene responsible for monolignol biosynthesis in rice culm[J]. Plant Cell Rep., 2012, 31(1): 91-101. |
| [1] | Man LIU, Yongxi HUANG, Pupu YAN, Jiali LIU, Liwei GUO. Identification of Shared Genetic Characteristics and Biological Mechanisms of Newcastle Disease and Streptococcal Disease in Chickens Based on the GEO Database [J]. Current Biotechnology, 2024, 14(3): 451-458. |
| [2] | Shi HUANG, Yinyin MO, Lyujing LUO, Huiting LIU, Zhengyu CHEN, Genliang LI. Bioinformatics Analysis on the Regulation Mechanism of NHP2 on Hepatocellular Carcinoma Senescence [J]. Current Biotechnology, 2024, 14(1): 141-148. |
| [3] | Xingshu ZHU, Junjie HUANG, Caiming WENG, Wangwu LIU, Yongyuan CHEN, Huiyan CHEN, Hu ZHAO, Yu WANG, Chengzhi LIN. Screening and Functional Enrichment Analysis of Key Hub Genes in Lynch Syndrome-associated Colorectal Cancer Based on Bioinformatics Analysis [J]. Current Biotechnology, 2023, 13(2): 291-297. |
| [4] | Fengmei LI, Xueyuan TANG, Gaoyang JIAO, Yuting QI, Xianting DU, Qianqian BAO, Meng ZHANG, Xiaobo XU, Xin XU, Yanfang ZHANG. Identification and Analysis of Genes Controlling Flower Shape in Lonicerajaponica Thunb. [J]. Current Biotechnology, 2023, 13(1): 90-101. |
| [5] | Min YU, Min WANG, Yanhuan WEI, Yiyi LIU. Analysis of Potential Key Molecular Biomarkers and Immune Infiltration Characteristics of SARS-CoV-2 Virus Infection [J]. Current Biotechnology, 2022, 12(5): 760-768. |
| [6] | XU Guangping, ZHANG Zhiyong, REN Zhonghong, ZHANG Zhiwei*. Bioinformatics Analysis of the Structure and Function of the Protein Encoded by ELO Gene in Acanthopagrus schlegelii [J]. Curr. Biotech., 2021, 11(3): 344-352. |
| [7] | XU Jie, WANG Kefei, WEI Xiaojing, GONG Lixin, JIAO Yang, QIU Lugui, HAO Mu*. Expression and Prognostic Significance of SCHIP1 in Acute Myeloid Leukemia: Analysis Based on Bioinformatics [J]. Curr. Biotech., 2020, 10(4): 417-425. |
| [8] | ZHANG Fengli1, YANG Yalin1*, XIA Rui1, GAO Chenchen1, LI Dong1, RAN Chao1, ZHANG Zhen2, ZHANG Hongling2, ZHOU Zhigang2*. Gene Cloning and Bioinformatics Analysis of Secret Protease Gene from Thelohanellus kitauei [J]. Curr. Biotech., 2019, 9(4): 375-383. |
| [9] | XU Changlu, ZHAO Yanhong, MA Yige, WANG Bingrui, WANG Ding, GUO Qing, TONG Jingyuan, GAO Jie, LI Yapu, LIU Jinhua, SHI Lihong*. Dissecting the Dynamic Expression of Autophagy Related Genes at Each Stage of Human Terminal Erythriod Differentiation Through Bioinformatic Methods [J]. Curr. Biotech., 2019, 9(3): 271-276. |
| [10] | SUN Jingjuan1, QIU Jingxuan1, ZENG Haijuan1, DING Chengchao1, WANG Guangbin2, LI Jie1, WANG Shujuan1, LIU Qing1*. Truncated Protein Expression, Purification and Polyclonal Antibody Preparation of PrsA1 Gene from Listeria monocytogenes [J]. Curr. Biotech., 2018, 8(3): 254-260. |
| [11] | JIANG Zhilei, ZHOU Lin, WU Fengci, SONG Xinyuan*. Analysis of Location and Expression Pattern of Maize Expansin Gene Family [J]. Curr. Biotech., 2018, 8(1): 34-40. |
| [12] | ZHANG Maona, JIANG Liang*, ZHANG Yan*. Advance on Bioinformatic Analysis of Selenium Metabolic Network and Selenoproteomes [J]. Curr. Biotech., 2017, 7(5): 537-544. |
| [13] | KANG Shuang1,2, HE Hong-xia2*, WANG Ming1*, GUO . The Acquisition and Bioinformatic Analysis of Sucrose Transporter Protein Gene PaSUC4 from Apricot [J]. Curr. Biotech., 2016, 6(2): 105-112. |
| [14] | LI Sheng-yan, LANG Zhi-hong*, HUANG Da-fang*. Research Progress on Eukaryotic Promoter [J]. Curr. Biotech., 2014, 4(3): 158-164. |
| [15] | YAO Zu-jie1, WU Song-qing1, PENG Yan1, YANG Mei2*. Biological Characteristics of Bacillus thuringiensis Strain BRC-PC6 and Cloning, Bioinformatics Analysis of its aiiA Gene [J]. Curr. Biotech., 2013, 3(2): 124-131. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||