Current Biotechnology ›› 2023, Vol. 13 ›› Issue (1): 90-101.DOI: 10.19586/j.2095-2341.2022.0071
• Articles • Previous Articles Next Articles
Identification and Analysis of Genes Controlling Flower Shape in Lonicerajaponica Thunb.
Fengmei LI( ), Xueyuan TANG, Gaoyang JIAO, Yuting QI, Xianting DU, Qianqian BAO, Meng ZHANG, Xiaobo XU, Xin XU, Yanfang ZHANG(
), Xueyuan TANG, Gaoyang JIAO, Yuting QI, Xianting DU, Qianqian BAO, Meng ZHANG, Xiaobo XU, Xin XU, Yanfang ZHANG( )
)
- School of Life Sciences & Basic Medicine,Xinxiang University,Henan Xinxiang 453003,China
-
Received:2022-05-09Accepted:2022-08-01Online:2023-01-25Published:2023-02-07 -
Contact:Yanfang ZHANG
金银花花形基因的发掘及生物信息学分析
李凤梅( ), 汤雪媛, 焦高洋, 齐玉亭, 杜献婷, 鲍倩倩, 张梦, 徐小博, 徐鑫, 张艳芳(
), 汤雪媛, 焦高洋, 齐玉亭, 杜献婷, 鲍倩倩, 张梦, 徐小博, 徐鑫, 张艳芳( )
)
- 新乡学院生命科学与基础医学学院,河南 新乡 453003
-
通讯作者:张艳芳 -
作者简介:李凤梅 E-mail: lfm3714167@163.com; -
基金资助:河南省高等学校重点科研项目计划项目(21B180009);2021年度新乡市科技攻关计划项目(GG2021014)
CLC Number:
Cite this article
Fengmei LI, Xueyuan TANG, Gaoyang JIAO, Yuting QI, Xianting DU, Qianqian BAO, Meng ZHANG, Xiaobo XU, Xin XU, Yanfang ZHANG. Identification and Analysis of Genes Controlling Flower Shape in Lonicerajaponica Thunb.[J]. Current Biotechnology, 2023, 13(1): 90-101.
李凤梅, 汤雪媛, 焦高洋, 齐玉亭, 杜献婷, 鲍倩倩, 张梦, 徐小博, 徐鑫, 张艳芳. 金银花花形基因的发掘及生物信息学分析[J]. 生物技术进展, 2023, 13(1): 90-101.
share this article
| 类型 | 调控部位 | 基因 |
|---|---|---|
| 其他调控花器官大小的因子 | ARGOS、OSR1、ANT、AILS、KLU、GRFs、GIFs、SWP、ARL、MED8、BB、DA1、DAR1、TCP4、MED25、BPEp/ARF8等 | |
| A类基因 | 花瓣、萼片 | AP1、AGL79、AP2、CLF、CAL、LUG、FUL、AGL79等 |
| B类基因 | 花瓣、雄蕊 | PI、APETALA3(AP3)、NAP、PIS、B类基因表达相关基因(LFY、UFO)等 |
| E类基因 | 花瓣 | SEP1、SEP2、SEP3、SEP4、AGL3等 |
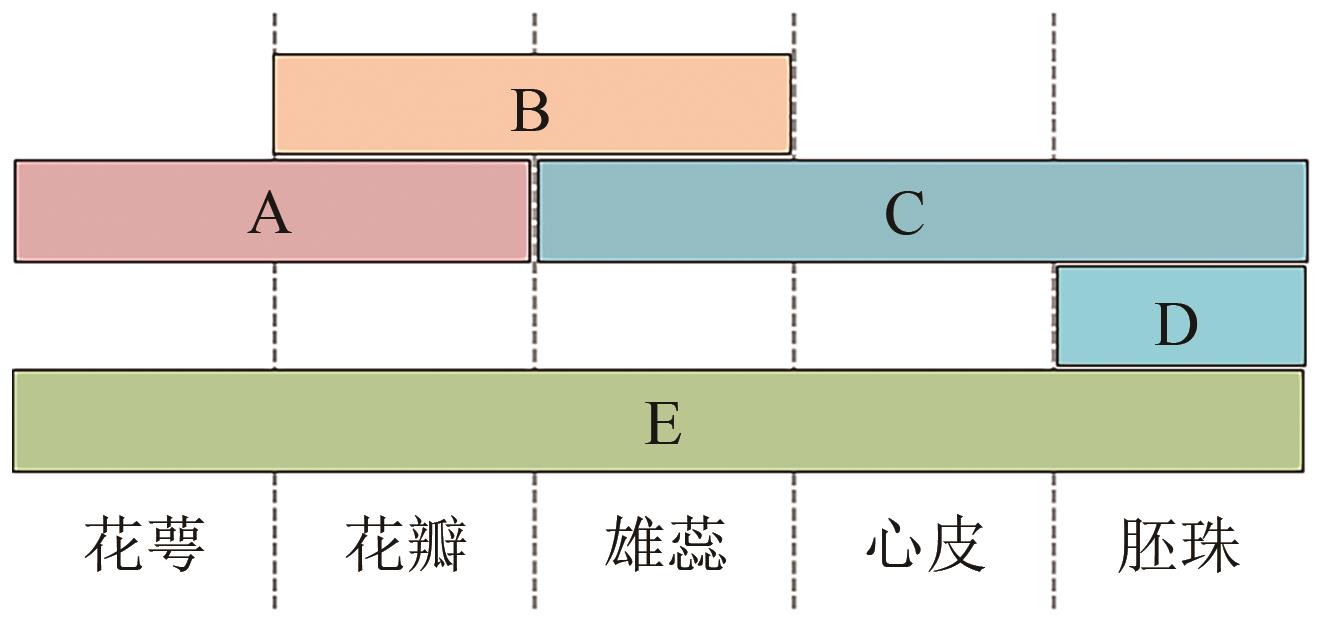
Table 1 Genes related to ABCDE model in Arabidopsis
| 类型 | 调控部位 | 基因 |
|---|---|---|
| 其他调控花器官大小的因子 | ARGOS、OSR1、ANT、AILS、KLU、GRFs、GIFs、SWP、ARL、MED8、BB、DA1、DAR1、TCP4、MED25、BPEp/ARF8等 | |
| A类基因 | 花瓣、萼片 | AP1、AGL79、AP2、CLF、CAL、LUG、FUL、AGL79等 |
| B类基因 | 花瓣、雄蕊 | PI、APETALA3(AP3)、NAP、PIS、B类基因表达相关基因(LFY、UFO)等 |
| E类基因 | 花瓣 | SEP1、SEP2、SEP3、SEP4、AGL3等 |
基因名称 (蛋白名称) | 染色体 | 位置 | 编码区 CDS | 正(+)反(-)向 | 结构域 | 拟南芥同源基因 (同一性) | |
|---|---|---|---|---|---|---|---|
同 一 基 因 簇 | GWHGAAZE001422 (GWHPAAZE001424) | 1 | 37616176~37616658 | 37616176~37616658 | - | MADS(1~61), low complexity(101~114), coiled coil(129~160) | AP1(40.000), AP3(34.177), SEP3(34.343) |
GWHGAAZE001424 (GWHPAAZE001426) | 1 | 37773949~37774431 | 37773949~37774431 | - | MADS(1~61), coiled coil(89~160), | AP1(38.667), AP3(34.177), SEP3(32.653), SEP4(47.158) | |
| 同一基因簇 | GWHGAAZE003600 (GWHPAAZE003603) | 1 | 100741719~100742564 | 100741719~100742564 | - | MADS(1~61), coiled coil(125~156), | AP1(47.059), AP3(37.500), SEP3(54.167) |
GWHGAAZE003601 (GWHPAAZE003604) | 1 | 100748839~100749663 | 100748839~100749663 | - | MADS(1~61) | AP1(46.154), AP3(38.889), SEP3(56.250), SEP4(54.167) | |
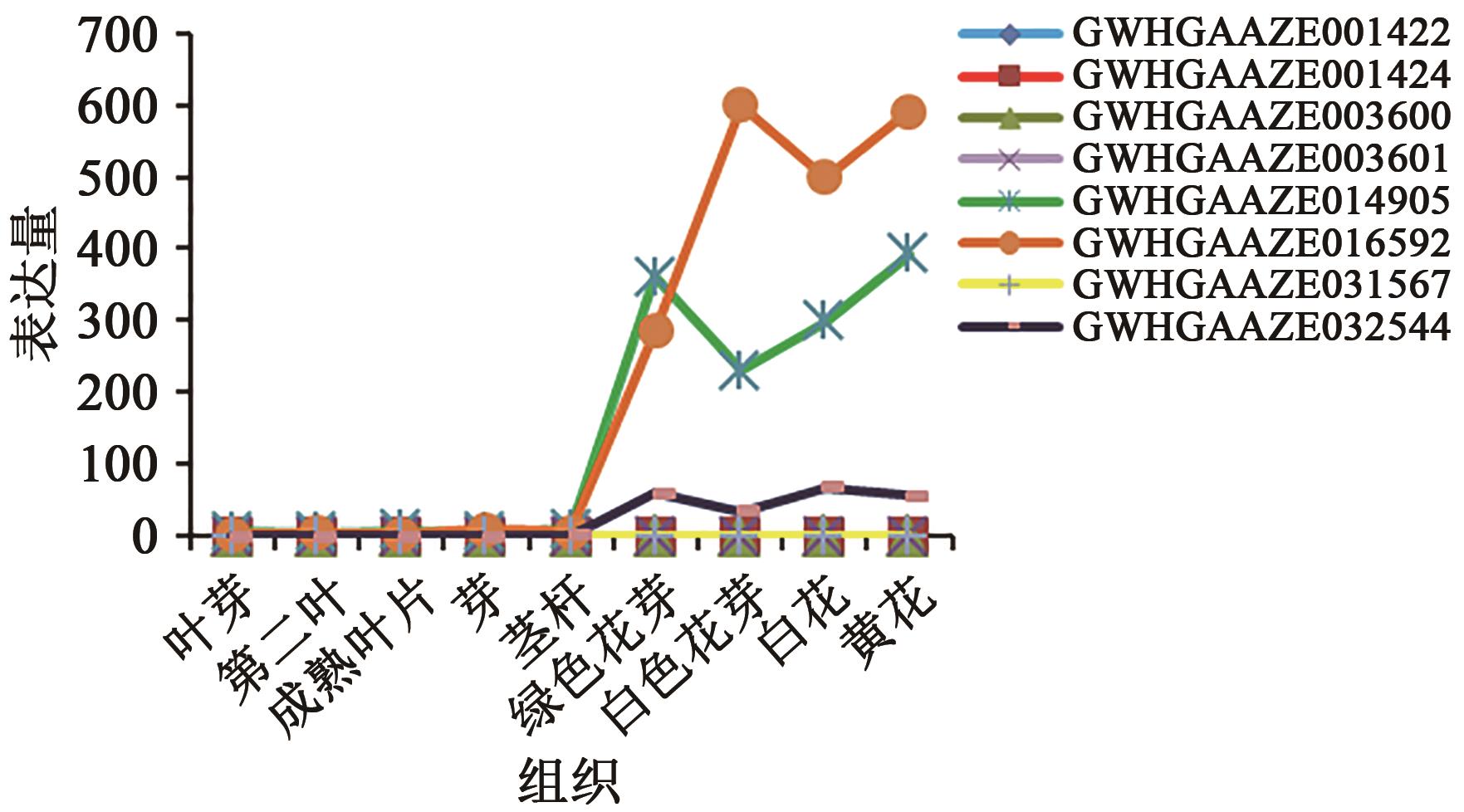
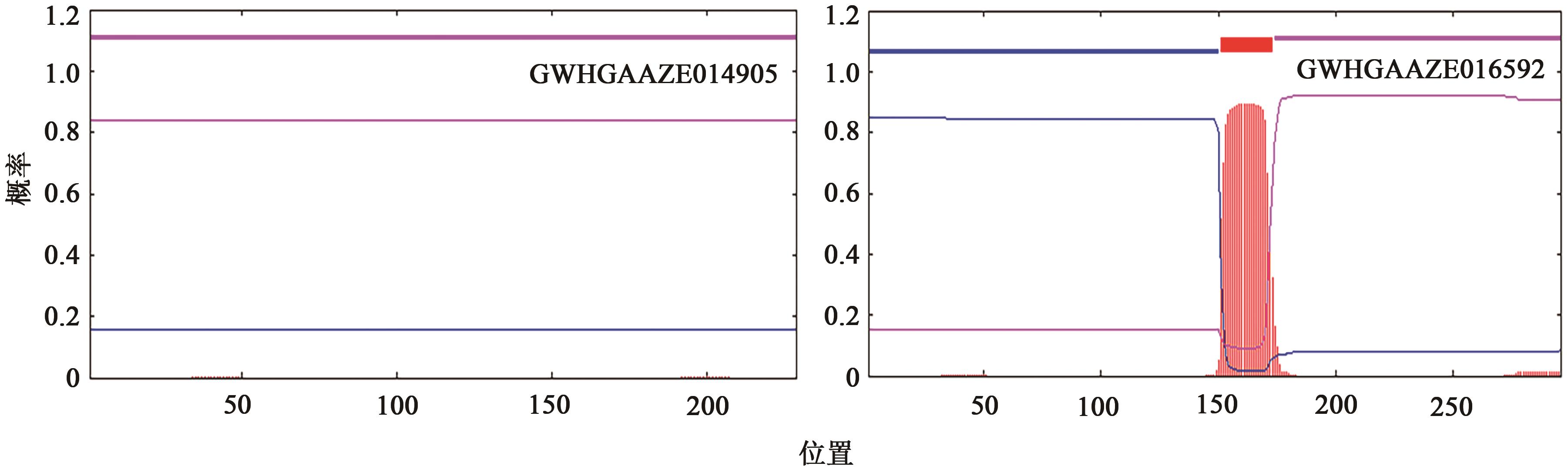
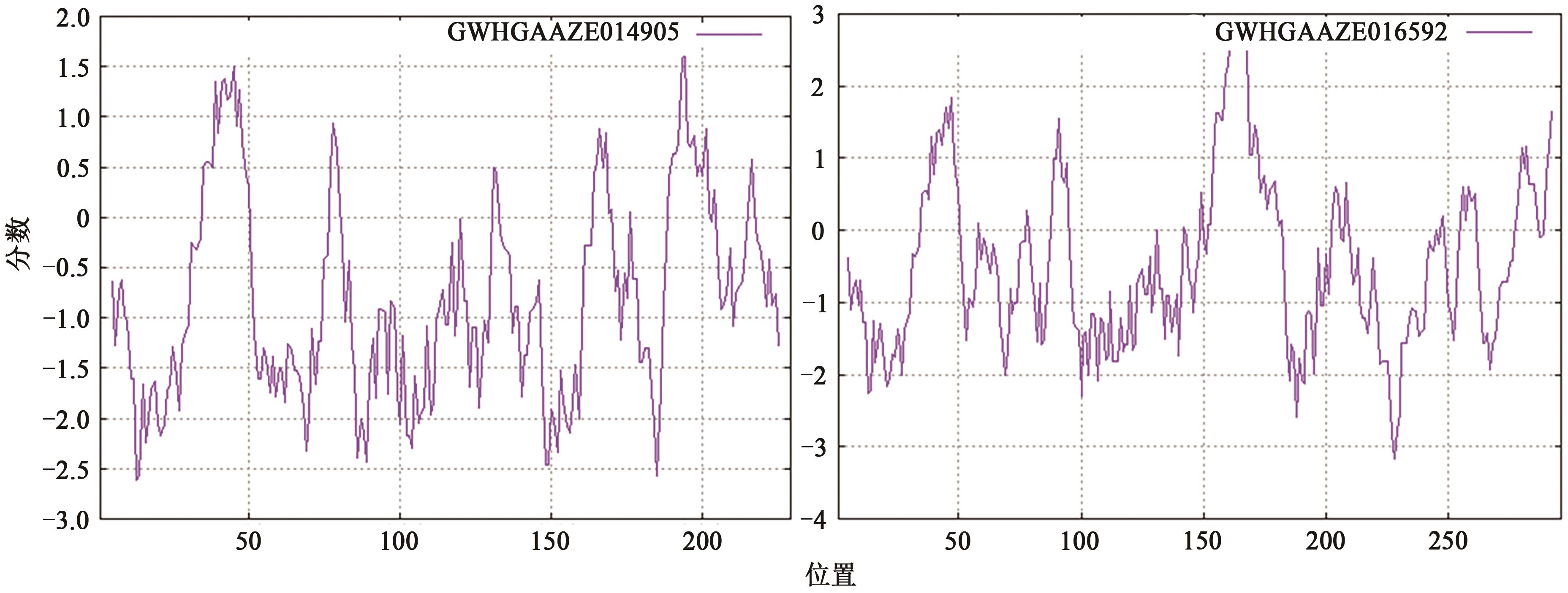
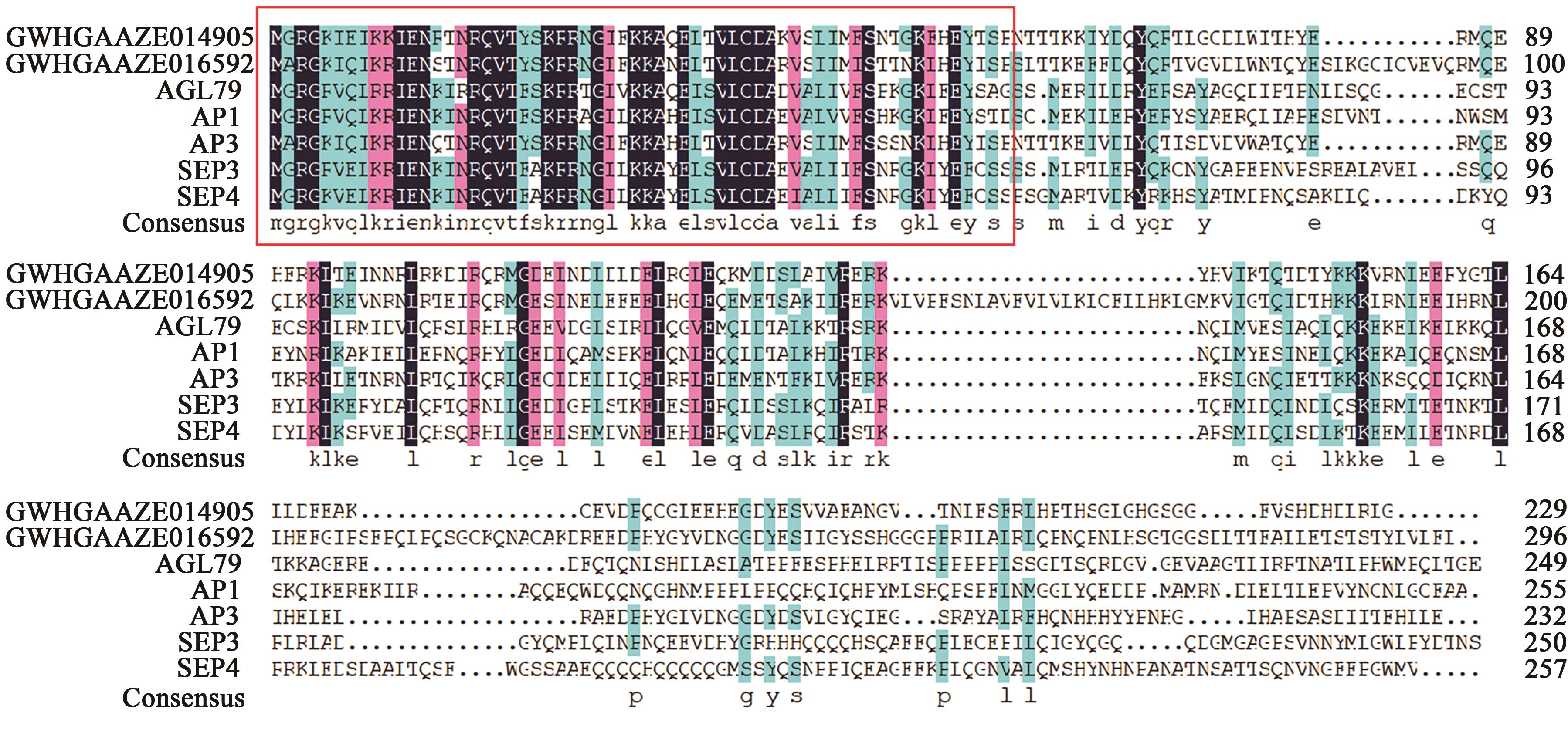
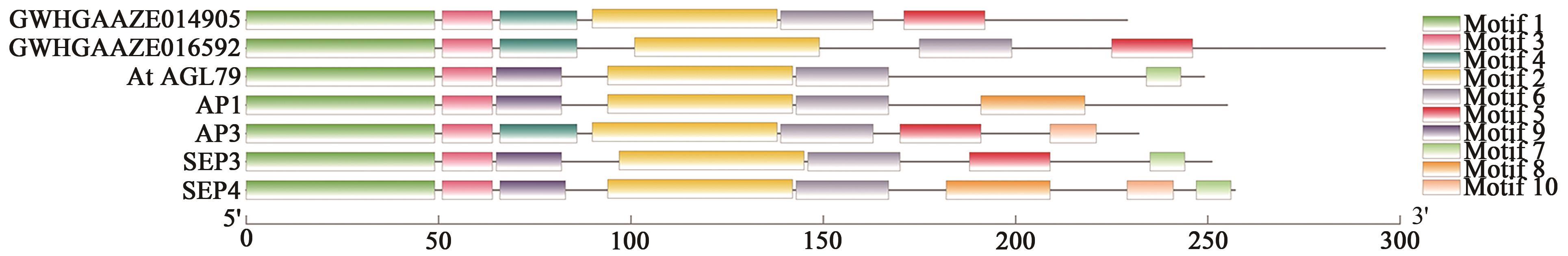
GWHGAAZE014905 (GWHPAAZE014912) | 4 | 21322982~21326145 | 21323004~21323191, 21324258~21324324, 21324428~21324489, 21324610~21324709, 21325160~21325201, 21325317~21325361, 21325548~21325733 | + | MADS(1~60), low complexity(111~121), coiled coil(144~164) | AP1(34.659), AP3(52.913), SEP3(35.859), SEP4(38.217) | |
GWHGAAZE016592 (GWHPAAZE016602) | 4 | 79255018~79260192 | 79255018~79255245, 79255659~79255718, 79257047~79257091, 79257197~79257238, 79258143~79258317, 79258437~79258531, 79259395~79259461, 79260005~79260192 | - | MADS(1~60), coiled coil(91~122), transmembrane region(151~173) | AGL79(37.662) AP1(38.272) AP3(50.345) SEP3(43.243) | |
GWHGAAZE031567 (GWHPAAZE031586) | 9 | 61028167~61028670 | 61028167~61028670 | - | MADS(1~60), coiled coil(78~117,140~167) | AP1(42.857) AP3(26.627) SEP3(45.833) | |
GWHGAAZE032544 (GWHPAAZE032564) | 21 | 4431033~4436189 | 4435923~4436104 4434534~4434609 4434055~4434122 4433812~4433911 4432855~4432896 4432353~4432394 4432092~4432216 4431316~4431415 | - | MADS(1~60), coiled coil(154~177) | AP1(42.286) AP3(36.559) SEP3(45.349) SEP4(39.429) | |
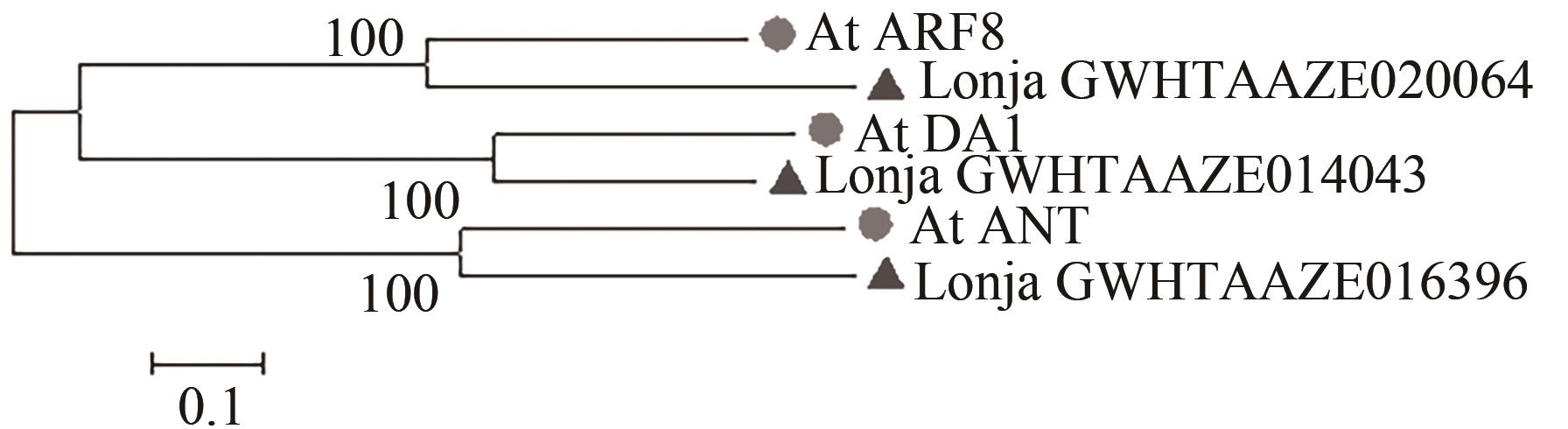
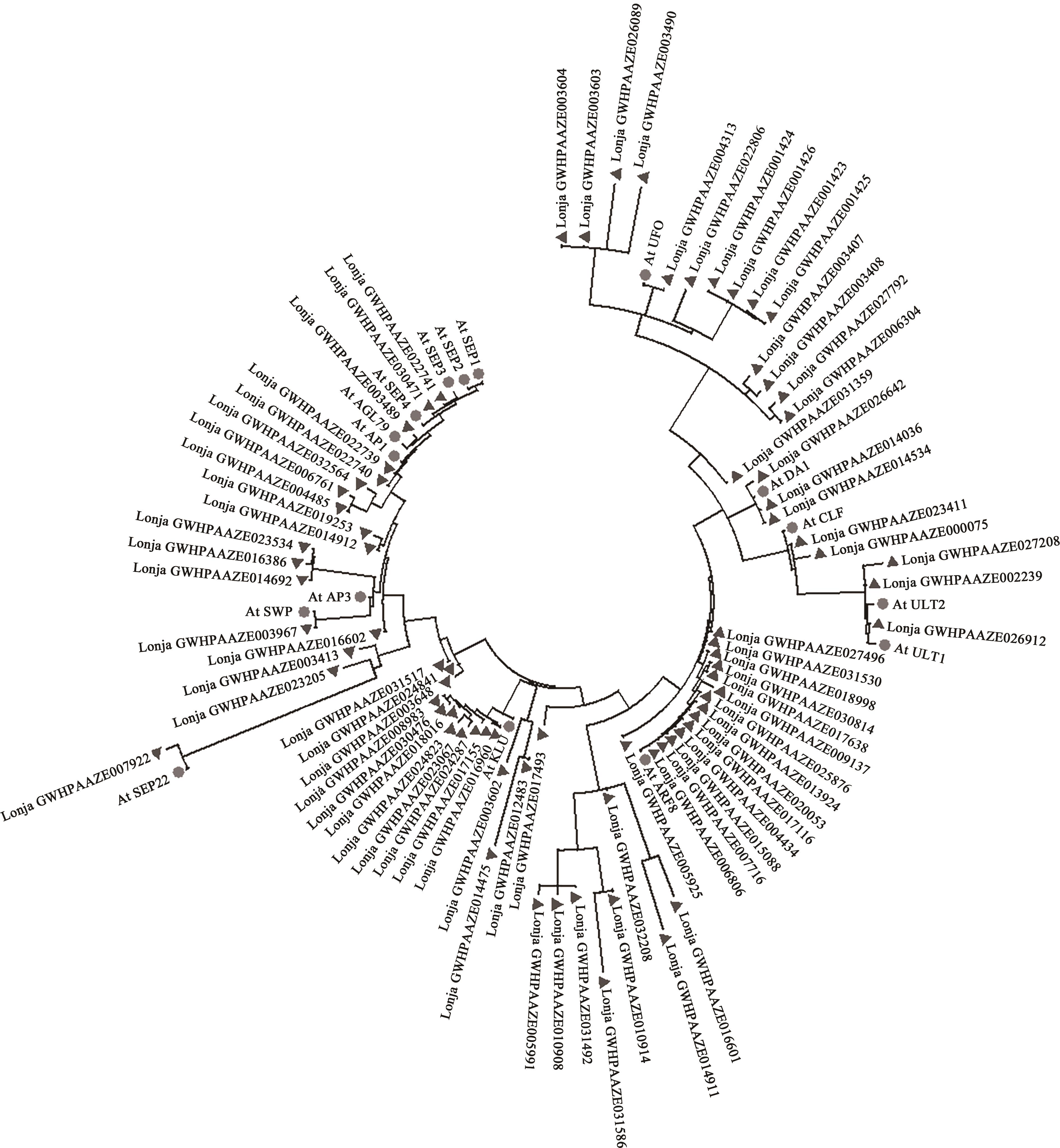
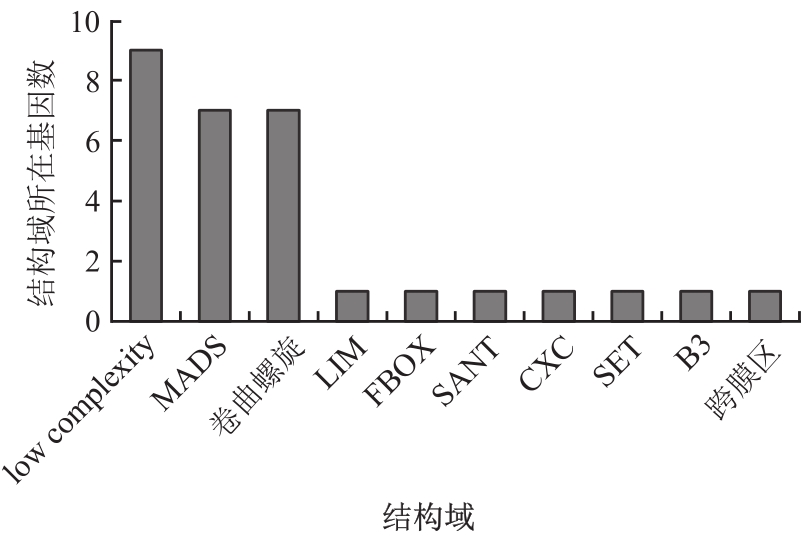
Table 2 Information list of candidate protein in Lonicera japonica Thunb.
基因名称 (蛋白名称) | 染色体 | 位置 | 编码区 CDS | 正(+)反(-)向 | 结构域 | 拟南芥同源基因 (同一性) | |
|---|---|---|---|---|---|---|---|
同 一 基 因 簇 | GWHGAAZE001422 (GWHPAAZE001424) | 1 | 37616176~37616658 | 37616176~37616658 | - | MADS(1~61), low complexity(101~114), coiled coil(129~160) | AP1(40.000), AP3(34.177), SEP3(34.343) |
GWHGAAZE001424 (GWHPAAZE001426) | 1 | 37773949~37774431 | 37773949~37774431 | - | MADS(1~61), coiled coil(89~160), | AP1(38.667), AP3(34.177), SEP3(32.653), SEP4(47.158) | |
| 同一基因簇 | GWHGAAZE003600 (GWHPAAZE003603) | 1 | 100741719~100742564 | 100741719~100742564 | - | MADS(1~61), coiled coil(125~156), | AP1(47.059), AP3(37.500), SEP3(54.167) |
GWHGAAZE003601 (GWHPAAZE003604) | 1 | 100748839~100749663 | 100748839~100749663 | - | MADS(1~61) | AP1(46.154), AP3(38.889), SEP3(56.250), SEP4(54.167) | |
GWHGAAZE014905 (GWHPAAZE014912) | 4 | 21322982~21326145 | 21323004~21323191, 21324258~21324324, 21324428~21324489, 21324610~21324709, 21325160~21325201, 21325317~21325361, 21325548~21325733 | + | MADS(1~60), low complexity(111~121), coiled coil(144~164) | AP1(34.659), AP3(52.913), SEP3(35.859), SEP4(38.217) | |
GWHGAAZE016592 (GWHPAAZE016602) | 4 | 79255018~79260192 | 79255018~79255245, 79255659~79255718, 79257047~79257091, 79257197~79257238, 79258143~79258317, 79258437~79258531, 79259395~79259461, 79260005~79260192 | - | MADS(1~60), coiled coil(91~122), transmembrane region(151~173) | AGL79(37.662) AP1(38.272) AP3(50.345) SEP3(43.243) | |
GWHGAAZE031567 (GWHPAAZE031586) | 9 | 61028167~61028670 | 61028167~61028670 | - | MADS(1~60), coiled coil(78~117,140~167) | AP1(42.857) AP3(26.627) SEP3(45.833) | |
GWHGAAZE032544 (GWHPAAZE032564) | 21 | 4431033~4436189 | 4435923~4436104 4434534~4434609 4434055~4434122 4433812~4433911 4432855~4432896 4432353~4432394 4432092~4432216 4431316~4431415 | - | MADS(1~60), coiled coil(154~177) | AP1(42.286) AP3(36.559) SEP3(45.349) SEP4(39.429) | |
| C1498H2415N425O444S12 | ||
| 296 | ||
Table 3 Physical properties and chemical analysis of candidate gene for flower shape
| C1498H2415N425O444S12 | ||
| 296 | ||
| 同源蛋白 | α螺旋 | 延伸链 | β转角 | 无规则卷曲 | 二级结构元件分布 |
|---|---|---|---|---|---|
| GWHGAAZE014905 | 56.33% | 13.10% | 8.30% | 22.27% |  |
| GWHGAAZE016592 | 44.26% | 20.61% | 6.42% | 28.72% |  |
| At AGL79 | 48.19% | 7.63% | 4.42% | 39.76% |  |
| AP1 | 57.65% | 7.45% | 2.75% | 32.16% |  |
| AP3 | 52.16% | 17.67% | 7.33% | 22.84% |  |
| SEP3 | 52.19% | 9.96% | 4.78% | 33.07% |  |
| SEP4 | 49.81% | 7.78% | 3.89% | 38.52% |  |
Table 4 Analysis of the secondary structure for candidate proteins
| 同源蛋白 | α螺旋 | 延伸链 | β转角 | 无规则卷曲 | 二级结构元件分布 |
|---|---|---|---|---|---|
| GWHGAAZE014905 | 56.33% | 13.10% | 8.30% | 22.27% |  |
| GWHGAAZE016592 | 44.26% | 20.61% | 6.42% | 28.72% |  |
| At AGL79 | 48.19% | 7.63% | 4.42% | 39.76% |  |
| AP1 | 57.65% | 7.45% | 2.75% | 32.16% |  |
| AP3 | 52.16% | 17.67% | 7.33% | 22.84% |  |
| SEP3 | 52.19% | 9.96% | 4.78% | 33.07% |  |
| SEP4 | 49.81% | 7.78% | 3.89% | 38.52% |  |
| 同源蛋白 | 模板 | 描述 | 同一性 | 覆盖度 | 范围 | |
|---|---|---|---|---|---|---|
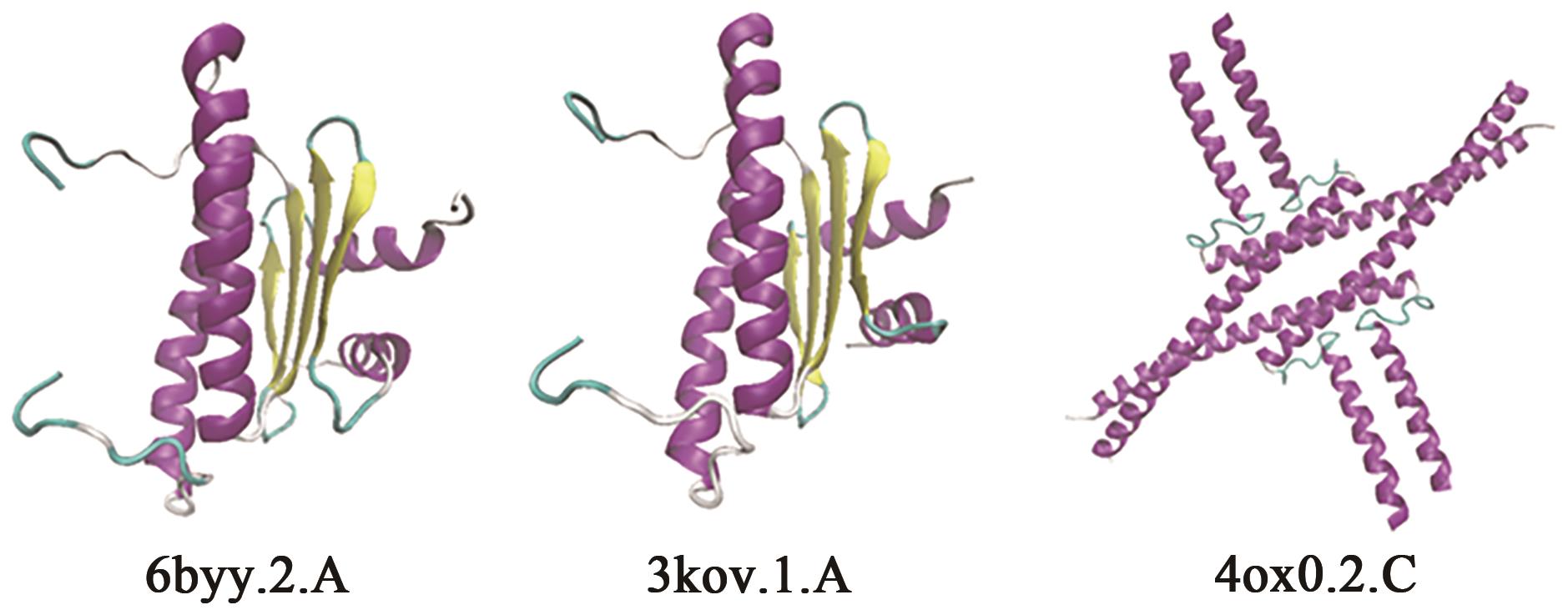
| GWHGAAZE014905 | 6byy.2.A | MEF2 CHIMERA/ MEF2 CHIMERA/DNA Complex | 42.67% | 0.33 | 2~77 |  |
| 4ox0.2.C | Developmental protein SEPALLATA 3/ Crystal structure of the keratin-like domain from the MADS transcription factor Sepallata 3 | 29.76% | 0.37 | 86~169 |  | |
| GWHGAAZE016592 | 6byy.2.A | 同上 | 44.59% | 0.25 | 2~76 |  |
| 4ox0.2.C | 同上 | 29.76% | 0.28 | 97~205 |  | |
| At AGL79 | 6byy.2.A | 同上 | 46.67% | 0.30 | 2~76 |  |
| 4ox0.2.C | 同上 | 36.47% | 0.34 | 90~174 |  | |
| AP1 | 3kov.1.A | Myocyte-specific enhancer factor 2A/ Structure of MEF2A bound to DNA reveals a completely folded MADS-box/MEF2 domain that recognizes DNA and recruits transcription co-factors | 56.34% | 0.28 | 2~74 |  |
| 4ox0.2.C | 同上 | 41.86% | 0.34 | 89~174 |  | |
| AP3 | 6byy.2.A | 同上 | 49.33% | 0.32 | 2~77 |  |
| 3-Sep | 6byy.2.A | 同上 | 48.24% | 0.34 | 2~88 |  |
| 4ox0.2.C | 同上 | 100.00% | 0.41 | 83~177 |  | |
| 4-Sep | 6byy.2.A | 同上 | 53.85% | 0.30 | 2~84 |  |
| 4ox0.2.C | 同上 | 54.65% | 0.33 | 89~174 |  | |
Table 5 Simulated 3D structure of candidate protein
| 同源蛋白 | 模板 | 描述 | 同一性 | 覆盖度 | 范围 | |
|---|---|---|---|---|---|---|
| GWHGAAZE014905 | 6byy.2.A | MEF2 CHIMERA/ MEF2 CHIMERA/DNA Complex | 42.67% | 0.33 | 2~77 |  |
| 4ox0.2.C | Developmental protein SEPALLATA 3/ Crystal structure of the keratin-like domain from the MADS transcription factor Sepallata 3 | 29.76% | 0.37 | 86~169 |  | |
| GWHGAAZE016592 | 6byy.2.A | 同上 | 44.59% | 0.25 | 2~76 |  |
| 4ox0.2.C | 同上 | 29.76% | 0.28 | 97~205 |  | |
| At AGL79 | 6byy.2.A | 同上 | 46.67% | 0.30 | 2~76 |  |
| 4ox0.2.C | 同上 | 36.47% | 0.34 | 90~174 |  | |
| AP1 | 3kov.1.A | Myocyte-specific enhancer factor 2A/ Structure of MEF2A bound to DNA reveals a completely folded MADS-box/MEF2 domain that recognizes DNA and recruits transcription co-factors | 56.34% | 0.28 | 2~74 |  |
| 4ox0.2.C | 同上 | 41.86% | 0.34 | 89~174 |  | |
| AP3 | 6byy.2.A | 同上 | 49.33% | 0.32 | 2~77 |  |
| 3-Sep | 6byy.2.A | 同上 | 48.24% | 0.34 | 2~88 |  |
| 4ox0.2.C | 同上 | 100.00% | 0.41 | 83~177 |  | |
| 4-Sep | 6byy.2.A | 同上 | 53.85% | 0.30 | 2~84 |  |
| 4ox0.2.C | 同上 | 54.65% | 0.33 | 89~174 |  | |
| 1 | WU J, WANG X, LIU Y, et al.. Flavone synthases from Lonicera japonica and L. macranthoides reveal differential flavone accumulation[J]. Sci. Rep., 2016, 6: 1-14. |
| 2 | WANG D, SHU X, ZHAO X, et al.. Comparative analysis of volatile oils from Lonicera japonica Thunb. var. chinensis wakey by HS-SPME and GC-MS[J]. Asian J. Chem., 2013, 25(2): 803-806. |
| 3 | ZHENG Z, ZHANG Q, CHEN R, et al.. Four new N-contained iridoid glycosides from flower buds of Lonicera japonica [J]. J. Asian Nat. Prod. Res., 2012, 14(8):729-737. |
| 4 | LIN L, ZHANG X, ZHU J, et al.. Two new triterpenoid saponins from the flowers and buds of Lonicera japonica [J]. J. Asian Nat. Prod. Res., 2008, 10(10): 925-929. |
| 5 | YAN L, QIU B, LI T, et al.. Green synthesis of silver nanoparticles from Lonicera japonica leaf extract and their anti-inflammatory and antibacterial effects[J]. Micro Nano. Lett., 2020,15(2): 90-95. |
| 6 | BANG B, PARK D, KWON K, et al.. BST-104, a water extract of Lonicera japonica, has a gastroprotective effect via antioxidant and anti-Inflammatory activities[J]. J. Med. Food, 2019, 22(4): 433-433. |
| 7 | LIU M, YU Q, YI Y, et al.. Antiviral activities of Lonicera japonica Thunb. components against grouper iridovirus in vitro and in vivo[J/OL]. Aquaculture, 2020, 519: 734882[2020-3-30]. . |
| 8 | ZHANG T, LIU H, BAI X, et al.. Fractionation and antioxidant activities of the water-soluble polysaccharides from Lonicera japonica Thunb.[J]. Int. J. Biol. Macromol., 2020, 151: 1058-1066. |
| 9 | YANG J, YU X, GUO L, et al.. Study of Lonicera japonica Thunb. against lung cancer based on pharmacological network[J]. Drug Eval. Res., 2020, 43 (2): 213-220. |
| 10 | ZHUANG J, DAI X, WU Q, et al.. A meta-analysis for Lianhua Qingwen on the treatment of coronavirus disease 2019 (COVID-19)[J/OL]. Complement. Ther. Med., 2021, 60: 102754[2021-6-19]. . |
| 11 | Chinese Pharmacopoeia Commission. Pharmacopeia of the People's Republic of China (中华人民共和国药典) [S]. Beijing: China Medical Science Press, 2020: 52, 230. |
| 12 | 孟雨婷,黄晓晨,侯元同,等. 花的形态与花发育的ABCDE模型[J]. 生物学杂志, 2017, 34(6): 105-115. |
| 13 | CAUSIERA B, SCHWARZ-SOMMERB Z, DAVIESA B. Floral organ identity: 20 years of ABCs[J]. Semin. Cell Dev. Biol., 2010, 21(1): 73-79. |
| 14 | PU X, LI Z, TINA Y, et al.. The honeysuckle genome provides insight into the molecular mechanism of carotenoid metabolism underlying dynamic flower coloration[J]. New Phytol., 2020, 227(3): 100-102. |
| 15 | XIAO Q, LI Z, QU M, et al.. LjaFGD: Lonicera japonica functional genomics database[J]. Integr. Plant Biol., 2021, 63(8): 1422-1436. |
| 16 | 林永强. 金银花药材及其制剂质量控制技术研究[D]. 济南: 山东大学, 2019. |
| 17 | MONDO M, KIMANI P, NARLA R. Validation of effectiveness marker-assisted gamete selection for multiple disease resistance in common bean[J]. Afr. Crop Sci. J., 2019, 27(4): 585-612. |
| 18 | O'BOYLE P, KELLY J, KIRK W. Use of marker-assisted selection to breed for resistance to common bacterial blight in common bean[J]. J. Am. Soc. Hortic. Sci., 2007, 132(3): 381-386. |
| 19 | HE L, XU X, LI Y, et al.. Transcriptome analysis of buds and leaves using 454 pyrosequencing to discover genes associated with the biosynthesis of active ingredients in Lonicera japonica Thunb[J/OL]. PLoS ONE, 2013, 8(4): 62922[2013-04-25]. . |
| 20 | FAN L, CHEN L, CUI W, et al.. Analysis of heavy metal content in edible honeysuckle (Lonicera japonica Thunb.) from China and health risk assessment[J]. J. Environ. Sci. Heal. B., 2020, 55(10):921-928. |
| 21 | 李建军,贾国伦,王君,等. 金银花不同种质花蕾的形态和品质成分比较分析[J]. 广西师范大学学报(自然科学版), 2013, 31(4) : 103-108. |
| 22 | 周连霞. 拟南芥CIB3调节成花转变的分子机制研究[D]. 长春: 吉林大学, 2021. |
| 23 | DENNIS L, PEACOCK J. Genes directing flower development in Arabidopsis [J]. Plant Cell, 2019, 31(6): 1192-1193. |
| 24 | REBOCHO A, KENNAWAY J, BANGHAM J, et al.. Formation and shaping of the antirrhinum flower through modulation of the CUP boundary gene[J]. Curr. Biol., 2017, 27(17): 2610-2622. |
| 25 | MUROYA M, OSHIMA H, KOBAYASHI S, et al.. Circadian clock in Arabidopsis thaliana determines flower opening time early in the morning and dominantly closes early in the afternoon[J]. Plant Cell Physiol., 2021, 62(5): 883-893. |
| 26 | BENEDITO V, VISSER P, VAN T J, et al.. Ectopic expression of LLAG1, an AGAMOUS homologue from lily (Lilium longiflorum Thunb.) causes floral homeotic modifications in Arabidopsis [J]. J. Exp. Bot., 2004, 55(401): 1391-1399. |
| 27 | GREGIS V, SESSA A, DORCA-FORNELL C, et al.. The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes[J]. Plant J., 2009, 60(4): 626-637. |
| 28 | RIECHMANN J, MEYEROWITZ E. Determination of floral organ identity by Arabidopsis MADS domain homeotic proteins AP1, AP3, PI, and AG is independent of their DNA-binding specificity[J]. Mol. Biol. Cell, 1997, 8 (7):1243-1259. |
| 29 | JING D, CHEN W, SHI M, et al.. Ectopic expression of an eriobotrya japonica APETALA3 ortholog rescues the petal and stamen identities in Arabidopsis ap3-3 mutant[J]. Biochem. Bioph. Res. Co., 2020, 523(1):33-38. |
| 30 | DITTA G, PINYOPICH A, ROBLES P, et al.. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity[J]. Curr. Biol., 2004, 14(21): 1935-1940. |
| 31 | FAVARO R, PINYOPICH A, BATTAGLIA R, et al.. MADS-box protein complexes control carpel and ovule development in Arabidopsis [J]. Plant Cell, 2003, 15(11):2603-2611. |
| 32 | 吕秀立. 栎属1考来木属等优良材料筛选和无性繁殖技术研究[D]. 南京: 南京林业大学, 2018. |
| 33 | FANG Z, QI R, LI X, et al.. Ectopic expression of FaesAP3, a Fagopyrum esculentum (Polygonaceae) AP3 orthologous gene rescues stamen development in an Arabidopsis ap3 mutant[J]. Gene, 2014, 550(2): 200-206. |
| [1] | XU Jie, WANG Kefei, WEI Xiaojing, GONG Lixin, JIAO Yang, QIU Lugui, HAO Mu*. Expression and Prognostic Significance of SCHIP1 in Acute Myeloid Leukemia: Analysis Based on Bioinformatics [J]. Curr. Biotech., 2020, 10(4): 417-425. |
| [2] | ZHANG Fengli1, YANG Yalin1*, XIA Rui1, GAO Chenchen1, LI Dong1, RAN Chao1, ZHANG Zhen2, ZHANG Hongling2, ZHOU Zhigang2*. Gene Cloning and Bioinformatics Analysis of Secret Protease Gene from Thelohanellus kitauei [J]. Curr. Biotech., 2019, 9(4): 375-383. |
| [3] | XU Changlu, ZHAO Yanhong, MA Yige, WANG Bingrui, WANG Ding, GUO Qing, TONG Jingyuan, GAO Jie, LI Yapu, LIU Jinhua, SHI Lihong*. Dissecting the Dynamic Expression of Autophagy Related Genes at Each Stage of Human Terminal Erythriod Differentiation Through Bioinformatic Methods [J]. Curr. Biotech., 2019, 9(3): 271-276. |
| [4] | YAO Zu-jie1, WU Song-qing1, PENG Yan1, YANG Mei2*. Biological Characteristics of Bacillus thuringiensis Strain BRC-PC6 and Cloning, Bioinformatics Analysis of its aiiA Gene [J]. Curr. Biotech., 2013, 3(2): 124-131. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||