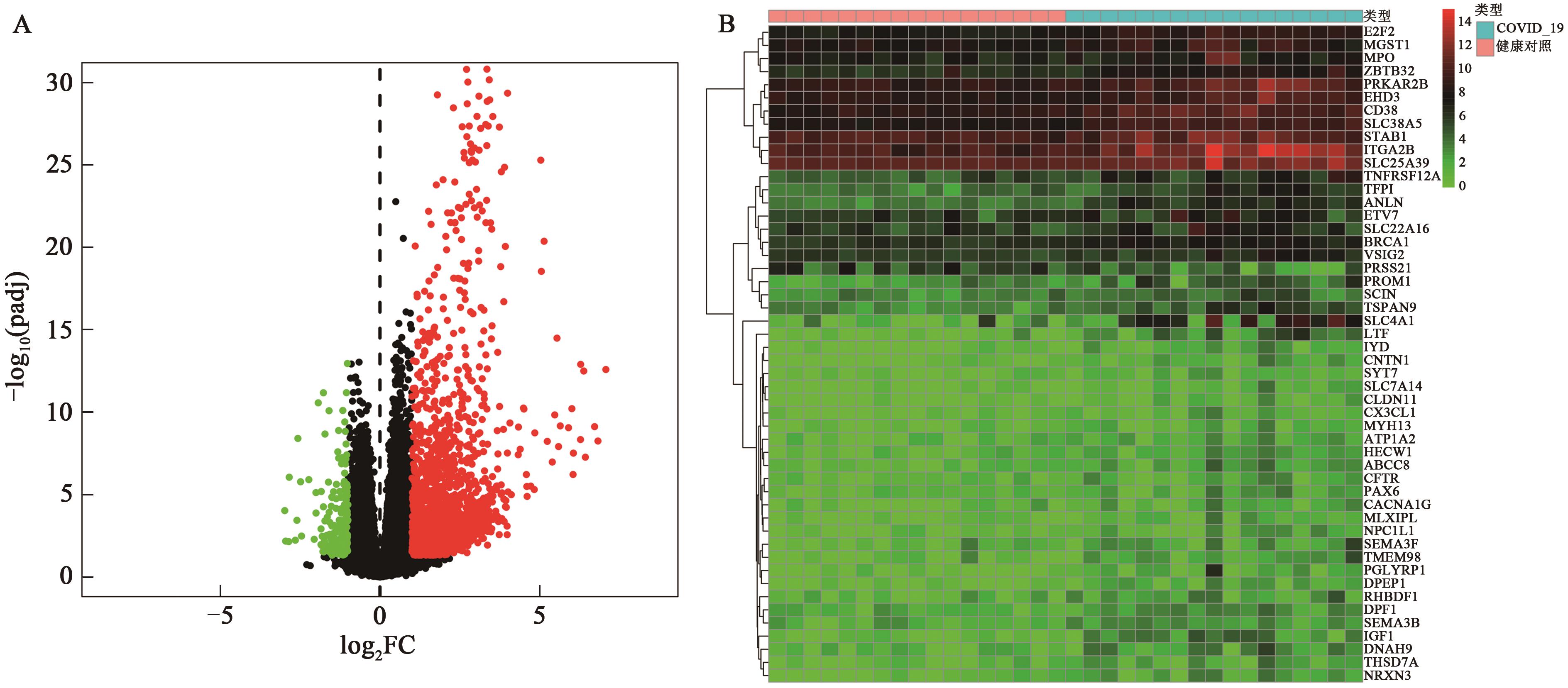
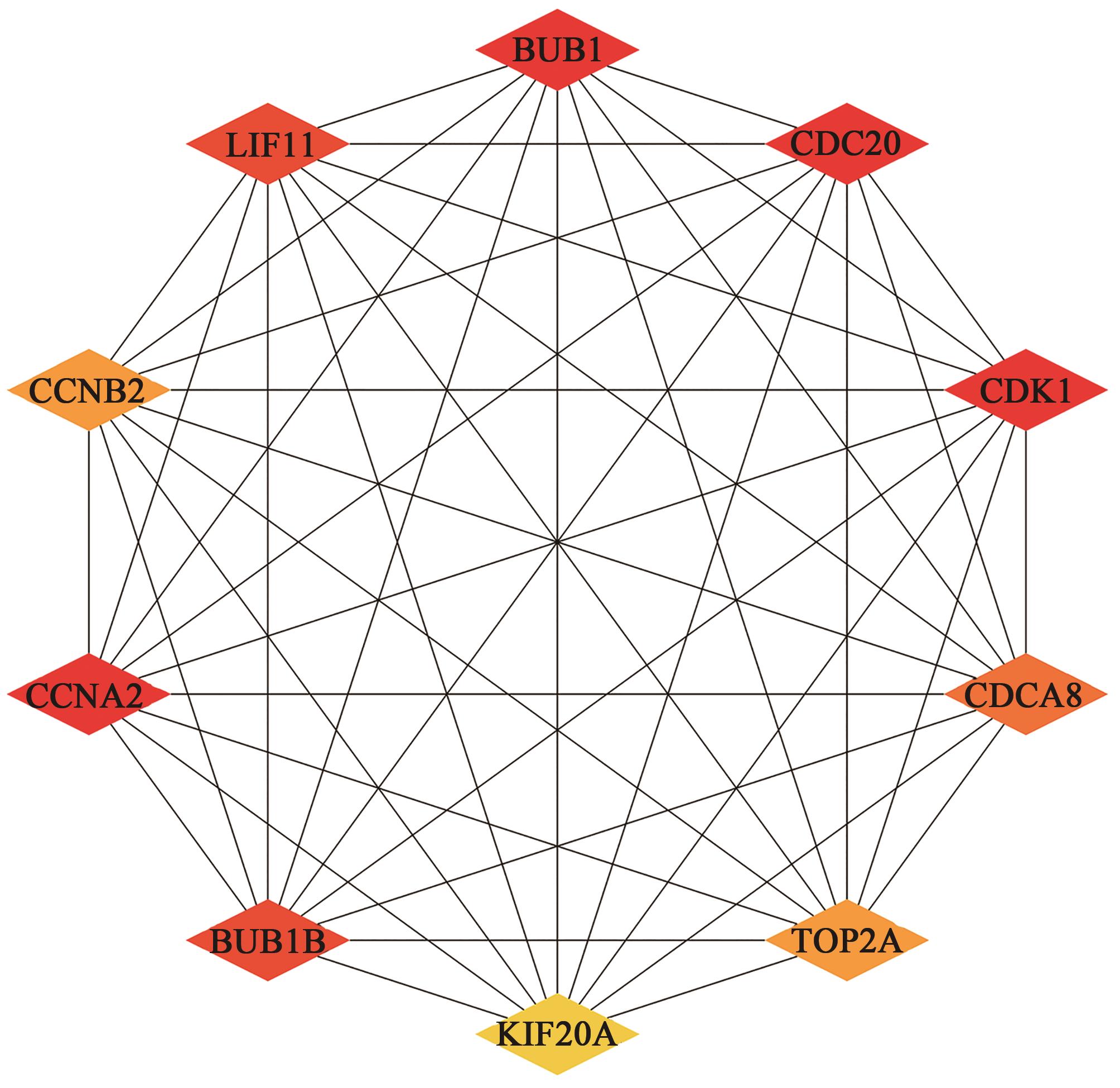
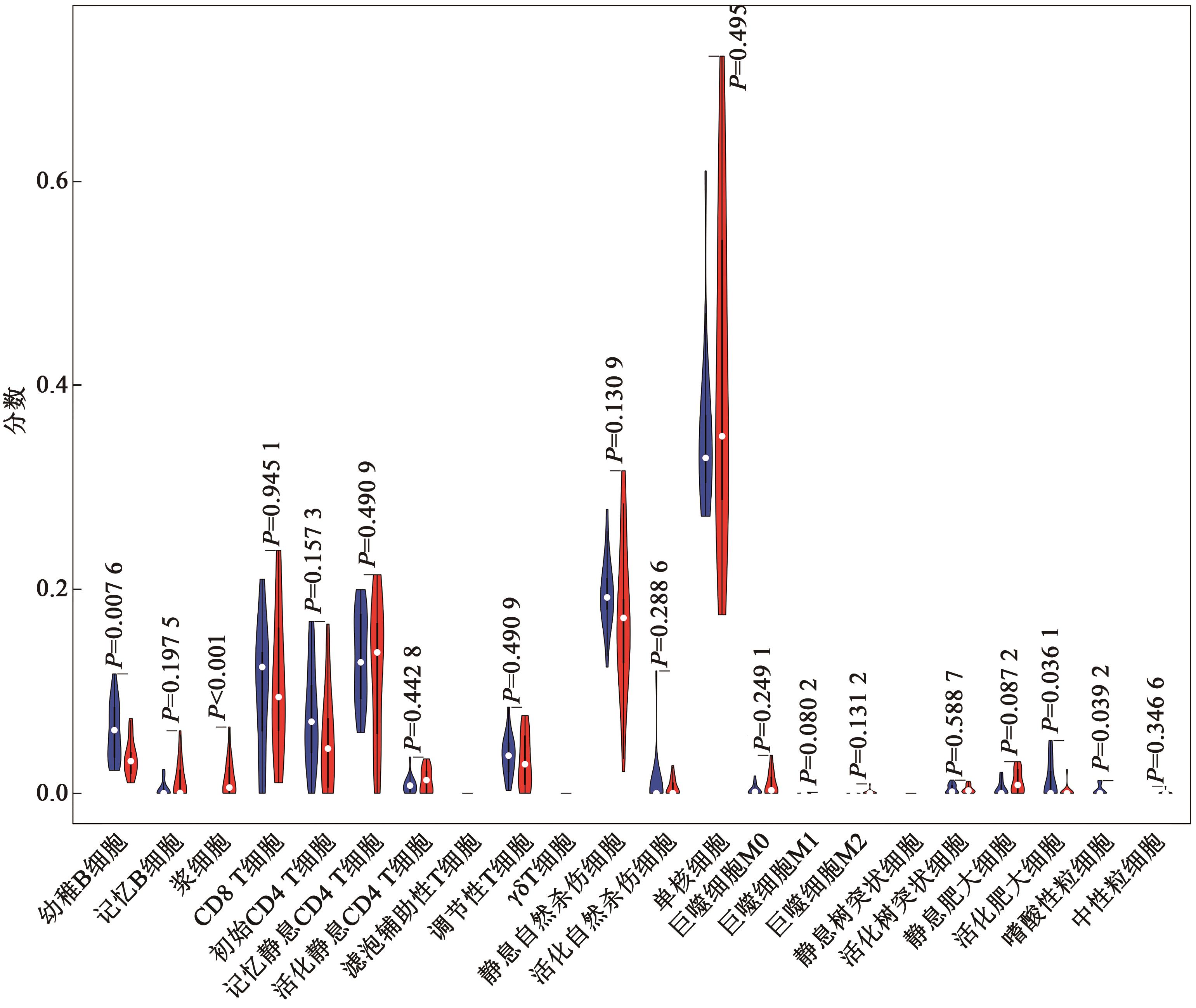
Current Biotechnology ›› 2022, Vol. 12 ›› Issue (5): 760-768.DOI: 10.19586/j.2095-2341.2022.0067
• Articles • Previous Articles Next Articles
Analysis of Potential Key Molecular Biomarkers and Immune Infiltration Characteristics of SARS-CoV-2 Virus Infection
Min YU1( ), Min WANG2, Yanhuan WEI1, Yiyi LIU1
), Min WANG2, Yanhuan WEI1, Yiyi LIU1
- 1.Department of Emergency Medicine,Rizhao People's Hospital,Shandong Rizhao 276800,China
2.Department of Laboratory Medicine,the Third Affiliated Hospital of Naval Military Medical University,Shanghai 200438,China
-
Received:2022-05-05Accepted:2022-06-30Online:2022-09-25Published:2022-09-30
SARS-CoV-2病毒感染潜在关键分子生物标志物及免疫浸润特征分析
- 1.日照市人民医院急诊医学科,山东 日照 276800
2.海军军医大学第三附属医院检验科,上海 200438
-
作者简介:于敏 E-mail:ym19912522707@163.com
CLC Number:
Cite this article
Min YU, Min WANG, Yanhuan WEI, Yiyi LIU. Analysis of Potential Key Molecular Biomarkers and Immune Infiltration Characteristics of SARS-CoV-2 Virus Infection[J]. Current Biotechnology, 2022, 12(5): 760-768.
于敏, 王敏, 魏延焕, 刘毅毅. SARS-CoV-2病毒感染潜在关键分子生物标志物及免疫浸润特征分析[J]. 生物技术进展, 2022, 12(5): 760-768.
share this article
| 分类 | ID | 描述 | 基因占比 | P值 | 基因数 |
|---|---|---|---|---|---|
| 生物学过程 | GO:0048285 | 细胞器裂变 | 61/632 | 6.88E-19 | 61 |
| GO:0000280 | 核分裂 | 60/632 | 1.57E-20 | 60 | |
| GO:0140014 | 有丝分裂核分裂 | 49/632 | 4.19E-21 | 49 | |
| GO:0007059 | 染色体分离 | 49/632 | 1.37E-17 | 49 | |
| GO:0044772 | 有丝分裂细胞周期相变 | 48/632 | 1.97E-13 | 48 | |
| 细胞组分 | GO:0098687 | 染色体区域 | 45/666 | 1.36E-14 | 45 |
| GO:0062023 | 含胶原细胞外基质 | 42/666 | 6.75E-10 | 42 | |
| GO:0000793 | 浓缩染色体 | 35/666 | 5.97E-13 | 35 | |
| GO:0005819 | 主轴 | 35/666 | 2.12E-07 | 35 | |
| GO:0000775 | 染色体,着丝粒区 | 34/666 | 5.88E-15 | 34 | |
| 分子功能 | GO:0046873 | 金属离子跨膜转运蛋白活性 | 28/651 | 0.001 538 | 28 |
| GO:0008017 | 微管结合 | 24/651 | 3.80E-05 | 24 | |
| GO:0005261 | 阳离子通道活性 | 24/651 | 0.000 911 | 24 | |
| GO:0005539 | 糖胺聚糖结合 | 20/651 | 0.000 210 | 20 | |
| GO:0015291 | 二级主动跨膜转运蛋白活性 | 20/651 | 0.000 368 | 20 |
Table 1 GO functional annotation and enrichment analysis
| 分类 | ID | 描述 | 基因占比 | P值 | 基因数 |
|---|---|---|---|---|---|
| 生物学过程 | GO:0048285 | 细胞器裂变 | 61/632 | 6.88E-19 | 61 |
| GO:0000280 | 核分裂 | 60/632 | 1.57E-20 | 60 | |
| GO:0140014 | 有丝分裂核分裂 | 49/632 | 4.19E-21 | 49 | |
| GO:0007059 | 染色体分离 | 49/632 | 1.37E-17 | 49 | |
| GO:0044772 | 有丝分裂细胞周期相变 | 48/632 | 1.97E-13 | 48 | |
| 细胞组分 | GO:0098687 | 染色体区域 | 45/666 | 1.36E-14 | 45 |
| GO:0062023 | 含胶原细胞外基质 | 42/666 | 6.75E-10 | 42 | |
| GO:0000793 | 浓缩染色体 | 35/666 | 5.97E-13 | 35 | |
| GO:0005819 | 主轴 | 35/666 | 2.12E-07 | 35 | |
| GO:0000775 | 染色体,着丝粒区 | 34/666 | 5.88E-15 | 34 | |
| 分子功能 | GO:0046873 | 金属离子跨膜转运蛋白活性 | 28/651 | 0.001 538 | 28 |
| GO:0008017 | 微管结合 | 24/651 | 3.80E-05 | 24 | |
| GO:0005261 | 阳离子通道活性 | 24/651 | 0.000 911 | 24 | |
| GO:0005539 | 糖胺聚糖结合 | 20/651 | 0.000 210 | 20 | |
| GO:0015291 | 二级主动跨膜转运蛋白活性 | 20/651 | 0.000 368 | 20 |
| ID | 描述 | 基因占比 | P值 | 基因数 |
|---|---|---|---|---|
| hsa04110 | 细胞周期 | 25/308 | 5.45E-12 | 25 |
| hsa04114 | 卵母细胞减数分裂 | 20/308 | 8.51E-08 | 20 |
| hsa04510 | 粘着斑 | 20/308 | 6.93E-05 | 20 |
| hsa05166 | 人类 T 细胞白血病病毒 Ⅰ型感染 | 18/308 | 0.001 840 | 18 |
| hsa04512 | ECM-受体相互作用 | 16/308 | 1.46E-07 | 16 |
| hsa04914 | 孕酮介导的卵母细胞成熟 | 15/308 | 5.86E-06 | 15 |
| hsa04261 | 心肌细胞中的肾上腺素信号传导 | 15/308 | 0.000 529 | 15 |
| hsa04218 | 细胞衰老 | 15/308 | 0.000 800 | 15 |
| hsa05410 | 肥厚型心肌病 | 13/308 | 3.00E-05 | 13 |
| hsa05414 | 扩张型心肌病 | 13/308 | 6.02E-05 | 13 |
Table 2 KEGG signaling pathway enrichment analysis
| ID | 描述 | 基因占比 | P值 | 基因数 |
|---|---|---|---|---|
| hsa04110 | 细胞周期 | 25/308 | 5.45E-12 | 25 |
| hsa04114 | 卵母细胞减数分裂 | 20/308 | 8.51E-08 | 20 |
| hsa04510 | 粘着斑 | 20/308 | 6.93E-05 | 20 |
| hsa05166 | 人类 T 细胞白血病病毒 Ⅰ型感染 | 18/308 | 0.001 840 | 18 |
| hsa04512 | ECM-受体相互作用 | 16/308 | 1.46E-07 | 16 |
| hsa04914 | 孕酮介导的卵母细胞成熟 | 15/308 | 5.86E-06 | 15 |
| hsa04261 | 心肌细胞中的肾上腺素信号传导 | 15/308 | 0.000 529 | 15 |
| hsa04218 | 细胞衰老 | 15/308 | 0.000 800 | 15 |
| hsa05410 | 肥厚型心肌病 | 13/308 | 3.00E-05 | 13 |
| hsa05414 | 扩张型心肌病 | 13/308 | 6.02E-05 | 13 |
| 1 | MARTIN B, DEWITT P E, RUSSELL S, et al.. Acute upper airway disease in children with the Omicron (B.1.1.529) variant of SARS-CoV-2-A report from the US national COVID cohort collaborative[J/OL]. JAMA Pediatr., 2022: e221110[2022-4-15]. . |
| 2 | CHAIYAKULSIL C, SRITIPSUKHO P, SATDHABUDHA A, et al.. An epidemiological study of pediatric COVID-19 in the era of the variant of concern[J/OL]. PLoS ONE, 2022, 17(4): e0267035[2022-4-15]. . |
| 3 | 杨镇州, 李妍, 杨雪, 等. 新型冠状病毒核酸检测试剂盒中酶的性能比较研究[J]. 生物技术进展, 2021, 11(6): 777-782. |
| 4 | BEERAKA N M, SUKOCHEVA O A, LUKINA E, et al.. Development of antibody resistance in emerging mutant strains of SARS CoV-2: Impediment for COVID-19 vaccines[J/OL]. Rev. Med. Virol., 2022, 13: e2346[2022-4-13]. . |
| 5 | MCLEAN G, KAMIL J, LEE B, et al.. The impact of evolving SARS-CoV-2 mutations and variants on COVID-19 vaccines[J/OL]. mBio, 2022, 13(2): e0297921[2022-4-26]. . |
| 6 | ZHANG C, FENG Y G, TAM C, et al.. Transcriptional profiling and machine learning unveil a concordant biosignature of Type I interferon-inducible host response across nasal swab and pulmonary tissue for COVID-19 diagnosis[J/OL]. Front Immunol., 2021, 12: 733171[2021-11-22]. . |
| 7 | ZHANG L, LI M, WANG Z, et al.. Cardiovascular risk after SARS-CoV-2 infection is mediated by IL18/IL18R1/HIF-1 signaling pathway axis[J/OL]. Front Immunol., 2022, 12: 780804[2022-1-5]. . |
| 8 | ARUNACHALAM P S, WIMMERS F, MOK C K P, et al.. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans[J]. Science, 2020, 369(6508): 1210-1220. |
| 9 | LOVE M I, HUBER W, ANDERS S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2[J/OL]. Genome Biol., 2014, 15(12): 550[2022-1-5]. . |
| 10 | LANGFELDER P, HORVATH S. WGCNA: an R package for weighted correlation network analysis[J/OL]. BMC Bioinform., 9:559[2008-12-29]. . |
| 11 | WU T, HU E, XU S, et al.. ClusterProfiler 4.0: A universal enrichment tool for interpreting omics data[J/OL]. Innovation (Camb), 2021, 2(3): 100141[2021-7-1]. . |
| 12 | NEWMAN A M, LIU C L, GREEN M R, et al.. Robust enumeration of cell subsets from tissue expression profiles[J]. Nat. Methods, 2015, 12(5): 453-457. |
| 13 | HU K. Become competent within one day in generating boxplots and violin plots for a novice without prior R experience[J/OL]. Methods Protoc., 2020, 3(4): 64[2020-9-23]. . |
| 14 | MELIDOU A, KÖDMÖN C, NAHAPETYAN K, et al.. Influenza returns with a season dominated by clade 3C.2a1b.2a.2 A(H3N2viruses), RegionWHOEuropean, 2021/22[J/OL]. Euro. Surveill., 2022, 27(15): 2200255[2022-4-14]. . |
| 15 | 刘家俊, 陈琛, 温明星, 等. 基于共表达网络和蛋白互作分析挖掘小麦赤霉病抗性相关核心蛋白[J]. 生物技术进展, 2021, 11(5): 628-633. |
| 16 | WANG X, BAI H, MA J, et al.. Identification of distinct immune cell subsets associated with asymptomatic infection, disease severity, and viral persistence in COVID-19 patients[J/OL]. Front Immunol., 2022, 13: 812514[2022-2-22]. . |
| 17 | HAHN F, HAMILTON S T, WANGEN C, et al.. Development of a PROTAC-based targeting strategy provides a mechanistically unique mode of anti-cytomegalovirus activity[J/OL]. Int. J. Mol. Sci., 2021, 22(23): 12858[2021-11-27]. . |
| 18 | AGRAWAL P, SAMBATURU N, OLGUN G, et al.. A path-based analysis of infected cell line and COVID-19 patient transcriptome reveals novel potential targets and drugs against SARS-CoV-2[J/OL]. Res. Squ., 2022, rs.3.rs-1474136[2022-3-21]. . |
| 19 | KIM Y J, WITWIT H, CUBITT B, et al.. Inhibitors of anti-apoptotic Bcl-2 family proteins exhibit potent and broad-spectrum anti-mammarenavirus activity via cell cycle arrest at G0/G1 phase[J/OL]. J. Virol., 2021, 95(24): e0139921[2021-11-23]. . |
| 20 | YUKA S, YILMAZ A. Effect of SARS-CoV-2 infection on host competing endogenous RNA and miRNA network[J/OL]. PeerJ, 2021, 9: e12370[2021-10-20]. . |
| 21 | AHMED F F, REZA M S, SARKER M S, et al.. Identification of host transcriptome-guided repurposable drugs for SARS-CoV-1 infections and their validation with SARS-CoV-2 infections by using the integrated bioinformatics approaches[J/OL]. PLoS ONE, 2022, 17(4): e0266124[2022-4-7]. . |
| 22 | CASCIOLA-ROSEN L, THIEMANN D R, ANDRADE F, et al.. IgM anti-ACE2 autoantibodies in severe COVID-19 activate complement and perturb vascular endothelial function[J/OL]. JCI Insight, 2022, 7(9): e158362[2022-5-9]. . |
| 23 | WILK A J, RUSTAGI A, ZHAO N Q, et al.. A single-cell atlas of the peripheral immune response in patients with severe COVID-19 [J]. Nat. Med., 2020, 26(7): 1070-1076. |
| 24 | KURI-CERVANTES L, PAMPENA M B, MENG W, et al.. Immunologic perturbations in severe COVID-19/SARS-CoV-2 infection[J/OL]. bioRxiv, 2020, 101717[2020-5-18]. . |
| 25 | ZHANG J J, DONG X, CAO Y Y, et al.. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China [J]. Allergy, 2020, 75(7): 1730-1741. |
| 26 | NAGASHIMA S, DUTRA A A, ARANTES M P, et al.. COVID-19 and lung mast cells: the kallikrein-kinin activation pathway[J/OL]. Int. J. Mol. Sci., 2022, 23(3): 1714[2022-2-2]. . |
| 27 | SUN Z, ZHANG Z, BANU K, et al.. Blood transcriptomes of SARS-CoV-2 infected kidney transplant recipients demonstrate immune insufficiency[J/OL]. MedRxiv, 2022: 22270203[2022-1-31]. . |
| [1] | Xiaoya LIU, Shuomin ZHANG, Peng ZHENG, Rui MA, Chaojun ZHANG. Role and Mechanisms of DBNDD1 in Colorectal Cancer Development [J]. Current Biotechnology, 2025, 15(4): 726-734. |
| [2] | Yaping HU, Zhenghong YU. Current Status of Research on mRNA Vaccines Against Viral Infectious Diseases [J]. Current Biotechnology, 2024, 14(4): 566-575. |
| [3] | Man LIU, Yongxi HUANG, Pupu YAN, Jiali LIU, Liwei GUO. Identification of Shared Genetic Characteristics and Biological Mechanisms of Newcastle Disease and Streptococcal Disease in Chickens Based on the GEO Database [J]. Current Biotechnology, 2024, 14(3): 451-458. |
| [4] | Liwen WANG, Jiangkun WANG, Bingbing WANG, Jianhong XU, Jianrong SHI, Xin LIU. Roles of Fusarium Toxins in Plant-pathogen Interaction [J]. Current Biotechnology, 2024, 14(2): 182-188. |
| [5] | Yeerkenbieke BUERLAN, Wenjia GUO, Xiaogang DONG. Advances on the Function of POSTN in Tumor Microenvironment [J]. Current Biotechnology, 2024, 14(2): 205-210. |
| [6] | Caihong WANG, Yuhan SHAO, Mengyuan JIANG. The Mechanisms of IL-17 Inhibiting Pseudomonas aeruginosa Infection on Lung Epithelial Cells [J]. Current Biotechnology, 2024, 14(2): 304-311. |
| [7] | Lili SUN, Yuemaierabola ANWAIER, Fuzhong LIU, Yeerkenbieke BUERLAN, Ye DILINAER, Wenjia GUO. Construction of Prognostic Prediction Model of Breast Cancer Based on Tumor-associated Fibroblast Genes and Analysis of Immune Infiltration [J]. Current Biotechnology, 2024, 14(2): 312-322. |
| [8] | Shi HUANG, Yinyin MO, Lyujing LUO, Huiting LIU, Zhengyu CHEN, Genliang LI. Bioinformatics Analysis on the Regulation Mechanism of NHP2 on Hepatocellular Carcinoma Senescence [J]. Current Biotechnology, 2024, 14(1): 141-148. |
| [9] | Changhu KE, Yaqing WU, Xueru DING, Huimin HUANG, Hui YAN. Research on the Mechanism of Jinyinhua Oral Liquid in the Prevention and Treatment of COVID-19 Based on Network Pharmacology and Molecular Docking [J]. Current Biotechnology, 2023, 13(5): 807-817. |
| [10] | Wenran HU, Xiaoyan HAO, Zhun ZHAO, Wukui SHAO, Shengqi GAO, Jianping LI, Quansheng HUANG. Bioinformatic Analysis of the CAD Gene Family in Cotton [J]. Current Biotechnology, 2023, 13(3): 412-424. |
| [11] | Qinqin LIU, Baiming HUANG, Yezi MA, Cuicui LIU, Hongtao WANG, Jiaxi ZHOU. Comparative Analysis of Megakaryocyte and Platelet Transcriptome Changes in Acute Infection [J]. Current Biotechnology, 2023, 13(3): 465-472. |
| [12] | Xingshu ZHU, Junjie HUANG, Caiming WENG, Wangwu LIU, Yongyuan CHEN, Huiyan CHEN, Hu ZHAO, Yu WANG, Chengzhi LIN. Screening and Functional Enrichment Analysis of Key Hub Genes in Lynch Syndrome-associated Colorectal Cancer Based on Bioinformatics Analysis [J]. Current Biotechnology, 2023, 13(2): 291-297. |
| [13] | Guangxuan TANG, Liwei HE, Yongyu CHEN, Jiayan ZHANG, Yaqi DAI, Huixiong LV, Lihui LIU. Techniques on Colonization and Detection of Plant Endophytes and its Application [J]. Current Biotechnology, 2023, 13(1): 55-64. |
| [14] | Minyi KAU, Xiao XU, Yiwen GAO, Ying CHEN, Zhengfei MA, Linxi YUAN. Narrative Review on the Roles of Selenium Against COVID-19 [J]. Current Biotechnology, 2023, 13(1): 72-76. |
| [15] | Fengmei LI, Xueyuan TANG, Gaoyang JIAO, Yuting QI, Xianting DU, Qianqian BAO, Meng ZHANG, Xiaobo XU, Xin XU, Yanfang ZHANG. Identification and Analysis of Genes Controlling Flower Shape in Lonicerajaponica Thunb. [J]. Current Biotechnology, 2023, 13(1): 90-101. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||