Current Biotechnology ›› 2021, Vol. 11 ›› Issue (4): 446-454.DOI: 10.19586/j.2095-2341.2021.0051
• Cutting-edge Technology • Previous Articles Next Articles
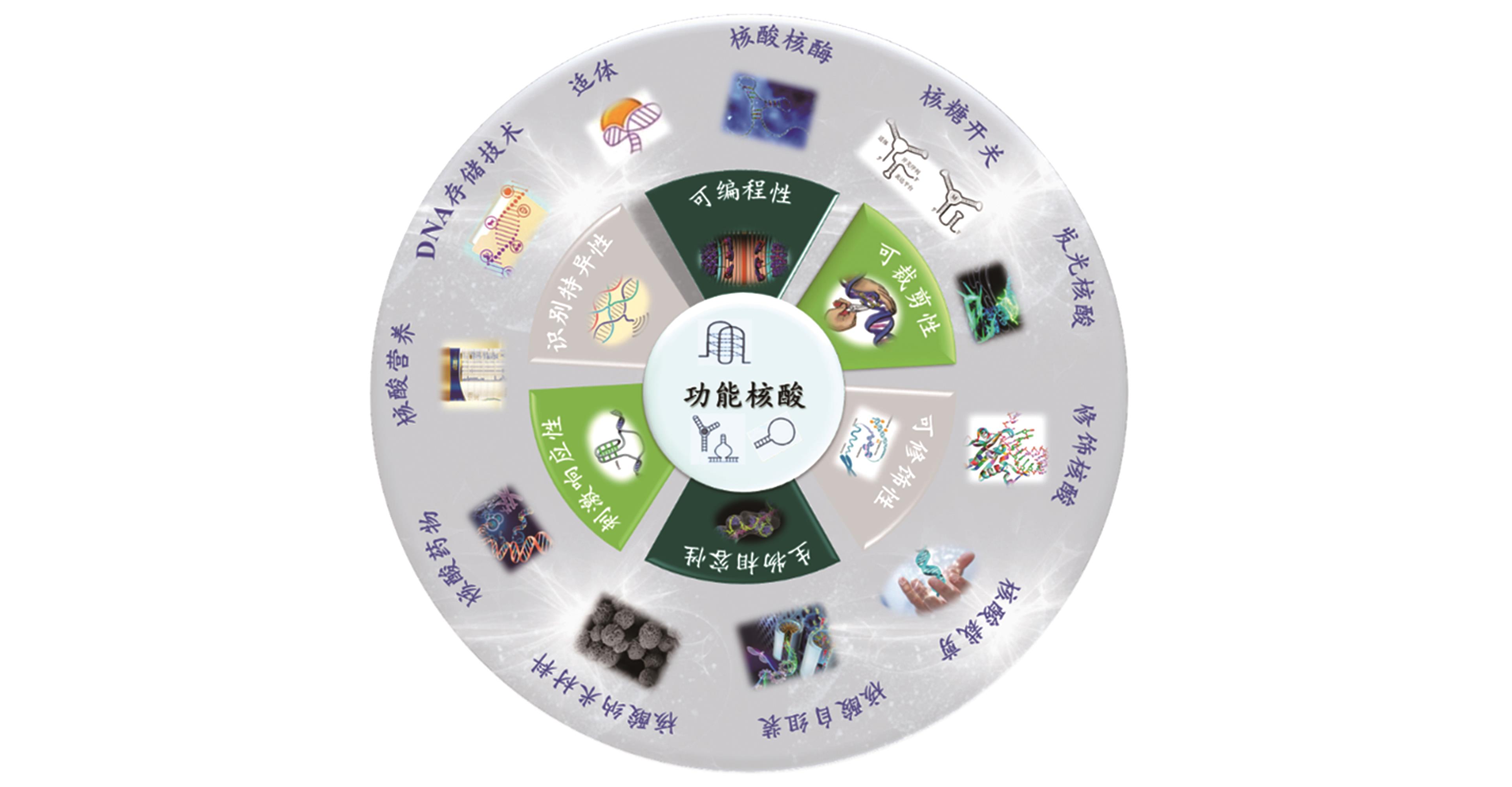
The Connotation and Extension of the Functional Nucleic Acid
Wentao XU( ), Min YANG, Longjiao ZHU, Yangzi ZHANG, Hongyu LI, Zaihui DU, Wenping YANG
), Min YANG, Longjiao ZHU, Yangzi ZHANG, Hongyu LI, Zaihui DU, Wenping YANG
- Key Laboratory of Precision Nutrition and Food Quality,Department of Nutrition and Health,China Agricultural University,Beijing 100083,China
-
Received:2021-04-10Accepted:2021-06-22Online:2021-07-25Published:2021-08-02
功能核酸概念的内涵与外延
许文涛( ), 杨敏, 朱龙佼, 张洋子, 李宏宇, 杜再慧, 杨文平
), 杨敏, 朱龙佼, 张洋子, 李宏宇, 杜再慧, 杨文平
- 中国农业大学营养与健康系,食品精准营养与质量控制教育部重点实验室,北京 100083
-
作者简介:许文涛E-mail:xuwentao@cau.edu.cn -
基金资助:山东省重大科技创新工程项目(2019JZZY011014);北京市农业科技项目(PXM2021_014207_000037)
CLC Number:
Cite this article
Wentao XU, Min YANG, Longjiao ZHU, Yangzi ZHANG, Hongyu LI, Zaihui DU, Wenping YANG. The Connotation and Extension of the Functional Nucleic Acid[J]. Current Biotechnology, 2021, 11(4): 446-454.
许文涛, 杨敏, 朱龙佼, 张洋子, 李宏宇, 杜再慧, 杨文平. 功能核酸概念的内涵与外延[J]. 生物技术进展, 2021, 11(4): 446-454.
share this article
| 类型 | 靶物质 | 作用位点 | 作用机制 | 总结 | 参考文献 |
|---|---|---|---|---|---|
| siRNA | mRNA | 细胞内(细胞质) | mRNA切割 | 根据RNAi的原理,具有与序列(siRNA)同源的mRNA切割的双链RNA,单链发夹RNA(shRNA)等 | [ |
| miRNA | microRNA | 细胞内(细胞质) | microRNA替代 | 双链RNA,单链发夹RNA的miRNA或其模拟物可用于增强因疾病而恶化的miRNA的功能 | [ |
反义核 苷酸 | mRNA、miRNA | 细胞内(细胞核及细胞质) | mRNA和miRNA 降解及拼接抑制 | 单链RNA/DNA,与靶mRNA和miRNA结合,引起降解或抑制,或在剪接时跳过外显子 | [ |
| 适体 | 蛋白质(胞外蛋白) | 细胞内(细胞核及细胞质) | 功能抑制 | 单链RNA/DNA,以与抗体/DNA相似的方式与靶蛋白结合 | [ |
寡聚核 苷酸 | 蛋白质(转录因子) | 细胞内(细胞核) | 转录抑制 | 与转录因子结合位点序列相同的双链DNA,其与受影响基因的转录因子结合以抑制靶基因 | [ |
| 核酸核酶 | RNA | 细胞内(细胞质) | RNA切割 | 具有酶功能的单链RNA,可结合和裂解靶RNA | [ |
| CpG oligo | 蛋白质(受体) | 细胞表面 | 免疫增强 | 具有CpG基序的寡脱氧核苷酸(单链DNA) | [ |
| 其他 | — | — | — | 除上面列出的核酸药物外,还可以激活先天免疫的核酸药物,例如PolyI:PolyC(双链RNA)和抗原 | [ |
Table 1 Classification summary of nucleic acid drug
| 类型 | 靶物质 | 作用位点 | 作用机制 | 总结 | 参考文献 |
|---|---|---|---|---|---|
| siRNA | mRNA | 细胞内(细胞质) | mRNA切割 | 根据RNAi的原理,具有与序列(siRNA)同源的mRNA切割的双链RNA,单链发夹RNA(shRNA)等 | [ |
| miRNA | microRNA | 细胞内(细胞质) | microRNA替代 | 双链RNA,单链发夹RNA的miRNA或其模拟物可用于增强因疾病而恶化的miRNA的功能 | [ |
反义核 苷酸 | mRNA、miRNA | 细胞内(细胞核及细胞质) | mRNA和miRNA 降解及拼接抑制 | 单链RNA/DNA,与靶mRNA和miRNA结合,引起降解或抑制,或在剪接时跳过外显子 | [ |
| 适体 | 蛋白质(胞外蛋白) | 细胞内(细胞核及细胞质) | 功能抑制 | 单链RNA/DNA,以与抗体/DNA相似的方式与靶蛋白结合 | [ |
寡聚核 苷酸 | 蛋白质(转录因子) | 细胞内(细胞核) | 转录抑制 | 与转录因子结合位点序列相同的双链DNA,其与受影响基因的转录因子结合以抑制靶基因 | [ |
| 核酸核酶 | RNA | 细胞内(细胞质) | RNA切割 | 具有酶功能的单链RNA,可结合和裂解靶RNA | [ |
| CpG oligo | 蛋白质(受体) | 细胞表面 | 免疫增强 | 具有CpG基序的寡脱氧核苷酸(单链DNA) | [ |
| 其他 | — | — | — | 除上面列出的核酸药物外,还可以激活先天免疫的核酸药物,例如PolyI:PolyC(双链RNA)和抗原 | [ |
| 1 | ELLINGTON A D, SZOSTAK J W. In vitro selection of RNA molecules that bind specific ligands [J]. Nature, 1990, 346(6287): 818-822. |
| 2 | TUERK C, GOLD L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase [J]. Science, 1990, 249(4968): 505-510. |
| 3 | DARMOSTUK M, RIMPELOVA S, GBELCOVA H, et al.. Current approaches in SELEX: An update to aptamer selection technology [J]. Biotechnol. Adv., 2015, 33(6): 1141-1161. |
| 4 | ABABNEH N, ALSHAER W, ALLOZI O, et al.. In vitro selection of modified RNA aptamers against CD44 cancer stem cell marker [J]. Nucl. Acid Ther., 2013, 23(6): 401-407. |
| 5 | BOSHTAM M, ASGARY S, KOUHPAYEH S, et al.. Aptamers against pro- and anti-inflammatory cytokines: a review [J]. Inflammation, 2017, 40(1): 340-349. |
| 6 | YERRAMILLI V S, KIM K H. Labeling RNAs in live cells using malachite green aptamer scaffolds as fluorescent probes [J]. ACS Synth. Biol., 2018, 7(3): 758-766. |
| 7 | WANG J, WANG Y, HU X, et al.. Dual-aptamer-conjugated molecular modulator for detecting bioactive metal ions and inhibiting metal-mediated protein aggregation [J]. Anal. Chem., 2019, 91(1): 823-829. |
| 8 | YAN A C, LEVY M. Aptamer-mediated delivery and cell-targeting aptamers: room for improvement [J]. Nucl. Acid Ther., 2018, 28(3): 194-199. |
| 9 | ABDELRASOUL G N, ANWAR A, MACKAY S, et al.. DNA aptamer-based non-faradaic impedance biosensor for detecting E. coli [J]. Anal. Chim. Acta, 2020, 1107: 135-144. |
| 10 | ZHANG Y, LAI B S, JUHAS M. Recent advances in aptamer discovery and applications [J]. Molecules, 2019, 24(5): 941-962. |
| 11 | ACHENBACH J, CHIUMAN W, CRUZ R, et al.. DNAzymes: from creation in vitro to application in vivo [J]. Curr. Pharm. Biotechnol., 2004, 5(4): 321-336. |
| 12 | ZAOURI N, CUI Z, PEINETTI A S, et al.. DNAzyme-based biosensor as a rapid and accurate verification tool to complement simultaneous enzyme-based media for E. coli detection [J]. Environ. Sci. Water Res. Technol., 2019, 5(12): 2260-2268. |
| 13 | ROTHENBROKER M, MCCONNELL E M, GU J, et al.. Selection and characterization of an RNA‐cleaving DNAzyme activated by legionella pneumophila [J]. Angew. Chem., 2021, 133(9): 4832-4838. |
| 14 | ZHANG S, LUAN Y, XIONG M, et al.. DNAzyme amplified aptasensing platform for ochratoxin A detection using a personal glucose meter [J]. ACS Appl. Mater. Interf., 2021, 13(8): 9472-9481. |
| 15 | NAHVI A, SUDARSAN N, EBERT M S, et al.. Genetic control by a metabolite binding mRNA [J]. Chem. Biol., 2002, 9(9): 1043-1049. |
| 16 | 沃森J D, 贝克T A,贝尔S P. 基因的分子生物学 [M]. 杨焕明译. 北京:科学出版社,2009. |
| 17 | 冯婧娴, 王佳稳, 林俊生, 等. 核酶核糖开关在基因治疗中的应用及挑战 [J]. 药学学报, 2014, 11: 1504-1511. |
| 18 | 毕茹茹, 范文廷, 马萍, 等. 核糖开关在细菌耐药性中的应用进展 [J]. 临床与病理杂志, 2016, 09(9): 1445-1449. |
| 19 | BEISEL C L, SMOLKE C D, ARKIN A P. Design principles for riboswitch function [J/OL]. PLoS Comp. Biol., 2009, 5(4): e1000363[2021-07-09]. . |
| 20 | 刘湛军, 路新枝, 姜鹏, 等. 核糖开关——药物开发的新靶点 [J]. 武汉大学学报(理学版), 2010, 56(3): 313-319. |
| 21 | 韩祥东, 刘薇, 吴存祥, 等. 核糖开关与基因表达调控 [J]. 中国生物化学与分子生物学报, 2011, 27(12): 1094-1100. |
| 22 | WANG X X, ZHU L J, LI S T, et al.. Fluorescent functional nucleic acid: principles, properties and applications in bioanalyzing [J]. Trac. Trend Anal. Chem., 2021, 116292-116325. |
| 23 | ZHOU Z, DING Y, SI S, et al.. Wide-field determination of aqueous mercury(II) based on tail-extensible DNA fluorescent probe with tunable dynamic range [J]. J. Hazard. Mater., 2021, 417:125975-125982. |
| 24 | TAN X, CONSTANTIN T P, SLOANE K L, et al.. Fluoromodules consisting of a promiscuous RNA aptamer and red or blue fluorogenic cyanine dyes: selection, characterization, and bioimaging [J]. J. Am. Chem. Soc., 2017, 139(26): 9001-9009. |
| 25 | DAVID P, CARDWELL L N, TAWIAH K D, et al.. Modular cell-internalizing aptamer nanostructure enables targeted delivery of large functional RNAs in cancer cell lines [J]. Nat. Commun., 2018, 9(1): 2283-2295. |
| 26 | LORENZ C, HADWIGER P, JOHN M, et al.. Steroid and lipid conjugates of siRNAs to enhance cellular uptake and gene silencing in liver cells [J]. Bioorg. Med. Chem. Lett., 2004, 14(19): 4975-4977. |
| 27 | NAIR J K, ATTARWALA H, SEHGAL A, et al.. Impact of enhanced metabolic stability on pharmacokinetics and pharmacodynamics of GalNAc-siRNA conjugates [J]. Nucl. Acids Res., 2017, 45(19): 10969-10977. |
| 28 | WAN W B, SETH P P. The medicinal chemistry of therapeutic oligonucleotides [J]. J. Med. Chem., 2016, 59(21): 9645-9667. |
| 29 | CHEN Z, LIU C, CAO F, et al.. DNA metallization: principles, methods, structures, and applications [J]. Chem. Soc. Rev., 2018, 47(11): 4017-4072. |
| 30 | YANG C, POHL R, TICHY M, et al.. Synthesis, photophysical properties, and biological profiling of benzothieno-fused 7-deazapurine ribonucleosides [J]. J. Org. Chem., 2020, 85(12): 8085-8101. |
| 31 | BING T, YANG X J, MEI H C, et al.. Conservative secondary structure motif of streptavidin-binding aptamers generated by different laboratories [J]. Bioorgan. Med. Chem., 2010, 18(5): 1798-1805. |
| 32 | PADLAN C S, MALASHKEVICH V N, ALMO S C, et al.. An RNA aptamer possessing a novel monovalent cation-mediated fold inhibits lysozyme catalysis by inhibiting the binding of long natural substrates [J]. RNA, 2014, 20(4): 447-461. |
| 33 | XIAO X N, ZHU L J, HE W C, et al.. Functional nucleic acids tailoring and its application [J]. Trac. Trend Anal. Chem., 2019, 118: 138-157. |
| 34 | LI M X, XU C H, ZHANG N, et al.. Exploration of the kinetics of toehold-mediated strand displacement via plasmon rulers [J]. ACS Nano, 2018, 12(4): 3341-3350. |
| 35 | WU C, CANSIZ S, ZHANG L, et al.. A nonenzymatic hairpin DNA cascade reaction provides high signal gain of mRNA imaging inside live cells [J]. J. Am. Chem. Soc., 2015, 137(15): 4900-4903. |
| 36 | YIN P, CHOI H M, CALVERT C R, et al.. Programming biomolecular self-assembly pathways [J]. Nature, 2008, 451(7176): 318-322. |
| 37 | RINKER S, KE Y, LIU Y, et al.. Self-assembled DNA nanostructures for distance-dependent multivalent ligand-protein binding [J]. Nat. Nanotechnol., 2008, 3(7): 418-422. |
| 38 | SEEMAN N C. Nucleic acid junctions and lattices [J]. J. Theor. Biol., 1982, 99(2): 237-247. |
| 39 | SEEMAN N C. DNA Nanotechnology: from the pub to information-based chemistry [J]. Methods Mol. Biol., 2018, 1811:1-9. |
| 40 | HU Y, NIEMEYER C M. From DNA nanotechnology to material systems engineering [J]. Adv. Mater., 2019, 31(26): e1806294[2021-07-09]. . |
| 41 | VYBORNYI M, VYBORNA Y, HANER R. DNA-inspired oligomers: from oligophosphates to functional materials [J]. Chem. Soc. Rev., 2019, 48(16): 4347-4360. |
| 42 | PINHEIRO A V, HAN D R, SHIH W M, et al.. Challenges and opportunities for structural DNA nanotechnology [J]. Nat. Nanotechnol., 2011, 6(12): 763-772. |
| 43 | KIM J, JANG D, PARK H, et al.. Functional-DNA-driven dynamic nanoconstructs for biomolecule capture and drug delivery [J]. Adv. Mater., 2018, 30(45): e1707351[2021-07-09]. . |
| 44 | XU X, WANG L, LI K, et al.. A smart DNA tweezer for detection of human telomerase activity [J]. Anal. Chem., 2018, 90(5): 3521-3530. |
| 45 | YURKE B, TURBERFIELD A J, MILLS A P, et al.. A DNA-fuelled molecular machine made of DNA [J]. Nature, 2000, 406(6796): 605-608. |
| 46 | ZHANG J, WANG L L, HOU M F, et al.. A ratiometric electrochemical biosensor for the exosomal microRNAs detection based on bipedal DNA walkers propelled by locked nucleic acid modified toehold mediate strand displacement reaction [J]. Biosens. Bioelect., 2018, 102: 33-40. |
| 47 | JUNG C, ALLEN P B, ELLINGTON A D. A simple, cleated DNA walker that hangs on to surfaces [J]. ACS Nano, 2017, 11(8): 8047-8054. |
| 48 | XUAN F, FAN T W, HSING I M. Electrochemical interrogation of kinetically-controlled dendritic DNA/PNA assembly for immobilization-free and enzyme-free nucleic acids sensing [J]. ACS Nano, 2015, 9(5): 5027-5033. |
| 49 | ANG Y S, YUNG L Y. Engineering self-contained DNA circuit for proximity recognition and localized signal amplification of target biomolecules [J]. Nucl. Acids Res., 2014, 42(14): 9523-9530. |
| 50 | XU W, HE W, DU Z, et al.. Functional nucleic acids‐nanomaterials: development, properties, and applications [J]. Angew. Chem. Intern. Edit., 2019, 60(13): 6890-6918. |
| 51 | 马翾, 张洋子, 许文涛. 功能核酸DNA水凝胶的制备与组装 [J]. 生物技术进展, 2019, 9(6): 554-562. |
| 52 | 马翾, 张洋子, 许文涛. 功能核酸DNA水凝胶的理化特性及应用进展 [J]. 生物技术进展, 2019, 9(6): 545-553. |
| 53 | ZHANG Y, ZHU L, TIAN J, et al.. Smart and functionalized development of nucleic acid-based hydrogels: assembly strategies, recent advances and challenges [J]. Adv. Sci., 2021, 2100216: 1-28. |
| 54 | LIU J. DNA-stabilized, fluorescent, metal nanoclusters for biosensor development [J]. Trends Anal. Chem., 2014, 58: 99-111. |
| 55 | XU J, ZHU X, ZHOU X, et al.. Recent advances in the bioanalytical and biomedical applications of DNA-templated silver nanoclusters [J]. Trends Anal. Chem., 2020, 124: 115786-115800. |
| 56 | SATYAVOLU N S R, LOH K Y, TAN L H, et al.. Discovery of and Insights into DNA "Codes" for Tunable Morphologies of Metal Nanoparticles [J]. Small, 2019, 15(26): e1900975[2021-07-12].. |
| 57 | O'NEILL P R, YOUNG K, SCHIFFELS D, et al.. Few-atom fluorescent silver clusters assemble at programmed sites on DNA nanotubes [J]. Nano Lett., 2012, 12(11): 5464-5469. |
| 58 | SU Y, CHU H, TIAN J, et al.. Insight into the nanomaterials enhancement mechanism of nucleic acid amplification reactions [J]. Trends Anal. Chem., 2021, 137: 116221-116237. |
| 59 | ZHOU W, FANG Y, REN J, et al.. DNA-templated silver and silver-based bimetallic clusters with remarkable and sequence-related catalytic activity toward 4-nitrophenol reduction [J]. Chem. Commun., 2019, 55(3): 373-376. |
| 60 | LU C, TANG L, GAO F, et al.. DNA-encoded bimetallic Au-Pt dumbbell nanozyme for high-performance detection and eradication of Escherichia coli O157:H7 [J]. Biosens. Bioelectr., 2021, 187: 113327-113336. |
| 61 | CHEN W, FANG X, YE X, et al.. Colorimetric DNA assay by exploiting the DNA-controlled peroxidase mimicking activity of mesoporous silica loaded with platinum nanoparticles [J]. Microchim. Acta, 2018, 185(12): 1-10. |
| 62 | A H M, WANG L, LI Y, et al.. Guanosine-rich aptamers Cu2O nanoparticles: enhanced peroxidase activity and specific recognition capability at neutral pH [J]. Chem. Commun., 2021,57, 643-646. |
| 63 | MENG Y, CHEN Y, ZHU J, et al.. Polarity control of DNA adsorption enabling the surface functionalization of CuO nanozymes for targeted tumor therapy [J]. Mater. Horizons, 2021, 8(3): 972-986. |
| 64 | BREAKER R R, JOYCE G F. The expanding view of RNA and DNA function [J]. Chem. Biol., 2014, 21(9): 1059-1065. |
| 65 | ZOU Y, SUN X, WANG Y, et al.. Single siRNA nanocapsules for effective siRNA brain delivery and glioblastoma treatment [J]. Adv. Mater., 2020, 32(24): e2000416[2021-07-12].. |
| 66 | CHAKRABORTY C, SHARMA A R, SHARMA G, et al.. Therapeutic miRNA and siRNA: moving from bench to clinic as next generation medicine [J]. Mol. Ther. Nucl. Acids, 2017, 8: 132-143. |
| 67 | LI D, MASTAGLIA F L, FLETCHER S, et al.. Precision medicine through antisense oligonucleotide-mediated exon skipping [J]. Trends Pharmacol. Sci., 2018, 39(11): 982-994. |
| 68 | JAIN S, KAUR J, PRASAD S, et al.. Nucleic acid therapeutics: a focus on the development of aptamers [J]. Expert Opin. Drug Discov., 2021, 16(3): 255-274. |
| 69 | MAHJOUBIN-TEHRAN M, REZAEI S, ATKIN S L, et al.. Decoys as potential therapeutic tools for diabetes [J/OL]. Drug Discov. Today, 2021, doi:10.1016/j.drudis.2021.04.004[2021-07-09]. . |
| 70 | KHAN A U, LAL S K. Ribozymes: a modern tool in medicine [J]. J. Biomed. Sci., 2003, 10(5): 457-467. |
| 71 | REPP L, RASOULIANBOROUJENI M, LEE H J, et al.. Acyl and oligo(lactic acid) prodrugs for PEG-b-PLA and PEG-b-PCL nano-assemblies for injection [J]. Offic. J. Contr. Release Soc., 2021, 330: 1004-1015. |
| 72 | CHARLEBOIS R, ALLARD B, ALLARD D, et al.. PolyI:C and CpG synergize with anti-ErbB2 mAb for treatment of breast tumors resistant to immune checkpoint inhibitors [J]. Cancer Res., 2017, 77(2): 312-319. |
| 73 | WRAIGHT C J, WHITE P J. Antisense oligonucleotides in cutaneous therapy [J]. Pharmacol. Therap., 2001, 90(1): 89-104. |
| 74 | CAVAGNARI B M. Gene therapy: nucleic acids as drugs. Action mechanisms and delivery into the cell] [J]. Archiv. Argent. Pediat., 2011, 109(3): 237-244. |
| 75 | 梁兴国, 李佥, 黄丽丽, 等. 核酸代谢与营养研究及发展趋势 [J]. 中国海洋大学学报(自然科学版), 2019, 10: 64-78. |
| 76 | KUBOTA A. Nutritional study of nucleotide components in the milk [J]. Nihon Shonika Gakkai Zasshi Acta Paed. Japon., 1969, 73(2):197-209. |
| 77 | EUN-HEE D, CHRISTOPHE C, SCHWAB C, et al.. Effect of dietary nucleosides and yeast extracts on composition and metabolic activity of infant gut microbiota in PolyFermS colonic fermentation models [J]. Fems Microbiol. Ecol., 2017,97(8):8[2021-07-12].. |
| 78 | XU M, LIANG R, GUO Q, et al.. Dietary nucleotides extend the life span in Sprague-Dawley rats[J]. J. Nutr. Health Aging, 2013, 17(3): 223-229. |
| 79 | OACUTE, PEZ-NAVARRO A T, GIL A, et al., Age-related effect of dietary nucleotides on liver nucleic acid content in rats [J]. Ann. Nutr. Metab., 1997, 41(5): 324-330. |
| 80 | CAI X, BAO L, WANG N, et al.. Dietary nucleotides protect against alcoholic liver injury by attenuating inflammation and regulating gut microbiota in rats [J]. Food Function, 2016, 7(6): 2898-2908. |
| 81 | CEZE L, NIVALA J, STRAUSS K. Molecular digital data storage using DNA[J]. Nat. Rev. Genet., 2019, 20: 456-466. |
| 82 | ORGANICK L, ANG S D, CHEN Y J, et al.. Random access in large-scale DNA data storage [J]. Nat. Biotechnol., 2018, 36(3): 242-250. |
| 83 | HECKEL R, MIKUTIS G, GRASS R N. A characterization of the DNA data storage channel [J]. Sci. Rep., 2019, 9(1): 1-12. |
| 84 | FARZADFARD F. DNA storage in everyday objects [J]. Nat. Biotechnol.,2020,38: 31-32. |
| 85 | LIM C K, NIRANTAR S, YEW W S,et al.. Novel modalities in DNA data storage[J/OL]. Trends Biotechnol., 2021,doi:10.1016/j.tibtech.2020.12.008[2021-07-09]. . |
| 86 | BANAL J L, SHEPHERD T R, BERLEANT J, et al.. Random access DNA memory using Boolean search in an archival file storage system[J/OL]. Nat. Mater, 2021,doi: 10.1038/s41563-021-01021-3[2021-07-09]. . |
| [1] | Yue SHI, Yao HAN, Hao LI, Yansong SUN. Application Progress of Biosensors Based on Field-effect Transistors in Nucleic Acid Detection [J]. Current Biotechnology, 2025, 15(4): 597-605. |
| [2] | Shuaishuai KANG, Ruian WANG, Wentao XU, Longjiao ZHU. Magnetic Aptamer Biosensors [J]. Current Biotechnology, 2023, 13(3): 339-344. |
| [3] | Enze CHENG-CHEN, Minghong JIA, Yueying LI. Research Progress of Nucleic Acid Reference Materials of Food-borne Pathogenic Bacteria [J]. Current Biotechnology, 2023, 13(2): 195-200. |
| [4] | Wenzhuo ZHAO, Chengxun LI, Zuojian HU, Hongxiu YU. Research Progress of Functional Nucleic Acid Used in Pathogenic Bacteria Detection [J]. Current Biotechnology, 2023, 13(1): 30-38. |
| [5] | Keren CHEN, Weishen WANG, Longjiao ZHU, Yangzi ZHANG, Xiaoyun HE, Kunlun HUANG, Wentao XU. Research Progress of Nucleic Acid⁃based Self⁃assembling Nanocarriers [J]. Current Biotechnology, 2022, 12(3): 352-357. |
| [6] | GERILEQIMUGE, NIU Zhenfeng, DONG Dan, ZHANG Taotao, ZHENG Rong. Application Progress of CRISPR-Cas System in Microbial Research [J]. Current Biotechnology, 2021, 11(3): 253-259. |
| [7] | LIU Wang, JIN Jinghao, CHEN Xiaoren. Application Progress of Loop-mediated Isothermal Amplification Technique [J]. Current Biotechnology, 2021, 11(2): 128-135. |
| [8] | YANG Jiayi1§*, CHEN Guifang2§, GAO Yunhuan1, WANG Zhidong1, WU Xiao1. Research Progress of Reference Material for HPV Nucleic Acid Detection [J]. Curr. Biotech., 2020, 10(6): 590-596. |
| [9] | HU Sihong, YOU Guoye. Application Prospects of Digital PCR in Detection of SARS-CoV-2 [J]. Curr. Biotech., 2020, 10(6): 674-679. |
| [10] | REN Wen1, YANG Haixia2, CHEN Lizhu2, LI Yufeng2, LIU Ya1*. Establishment and Application of Nucleic Acid Chromatography for Rapid Detection of Transgenic Plants [J]. Curr. Biotech., 2020, 10(6): 680-687. |
| [11] | XU Lei1,XIAO Guiqing2,SHENG Xiaojing1,QI Zhiqing1*,DIAO Yong1*. Research Progress on PCR Enhancer in Nucleic Acid in vitro Amplification Detection Technology [J]. Curr. Biotech., 2020, 10(2): 137-143. |
| [12] | MA Xuan1, ZHANG Yangzi1, XU Wentao1,2*. Progress on Physicochemical Properties and Applications of Functional Nucleic Acid DNA Hydrogels [J]. Curr. Biotech., 2019, 9(6): 545-553. |
| [13] | MA Xuan1, ZHANG Yangzi1, XU Wentao1,2*. Preparation and Assembly of Functional Nucleic Acid DNA Hydrogels [J]. Curr. Biotech., 2019, 9(6): 554-562. |
| [14] | YANG Wenping1,2, WU Yuangen1*. Colorimetric Sensors for Cd2+ and Pb2+ Detection Based on the Peroxidase Activity of Fe3O4 [J]. Curr. Biotech., 2019, 9(6): 611-619. |
| [15] | PENG Zhi1, CHEN Gangyi2, LIU Xuefei1, HUANG Xinhe1*. Application of Isothermal Nucleic Acid Amplification Technique in Pathogen Detection [J]. Curr. Biotech., 2018, 8(4): 284-292. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||