Current Biotechnology ›› 2024, Vol. 14 ›› Issue (3): 433-441.DOI: 10.19586/j.2095-2341.2023.0162
• Articles • Previous Articles Next Articles
Study on the Involvement of Auxin Signaling Pathway in Response to Powdery Mildew Stress in Pumpkin
Hui ZHANG1( ), Bobo LIU1, Fengmei LI1(
), Bobo LIU1, Fengmei LI1( ), Jian CUI2(
), Jian CUI2( )
)
- 1.College of Marine Science and Biological Engineering,Qingdao University of Science & Technology,Shandong Qingdao 266042,China
2.Qingdao Institute of Agricultural Science Research,Shandong Qingdao 266199,China
-
Received:2023-12-14Accepted:2024-01-24Online:2024-05-25Published:2024-06-18 -
Contact:Fengmei LI,Jian CUI
生长素信号途径参与南瓜白粉菌胁迫的响应研究
- 1.青岛科技大学海洋科学与生物工程学院,山东 青岛 266042
2.青岛市农业科学研究院,山东 青岛 266199
-
通讯作者:李凤梅,崔健 -
作者简介:张慧 E-mail: 824572941@qq.com; -
基金资助:国家自然科学基金项目(32072654)
CLC Number:
Cite this article
Hui ZHANG, Bobo LIU, Fengmei LI, Jian CUI. Study on the Involvement of Auxin Signaling Pathway in Response to Powdery Mildew Stress in Pumpkin[J]. Current Biotechnology, 2024, 14(3): 433-441.
张慧, 刘愽愽, 李凤梅, 崔健. 生长素信号途径参与南瓜白粉菌胁迫的响应研究[J]. 生物技术进展, 2024, 14(3): 433-441.
share this article
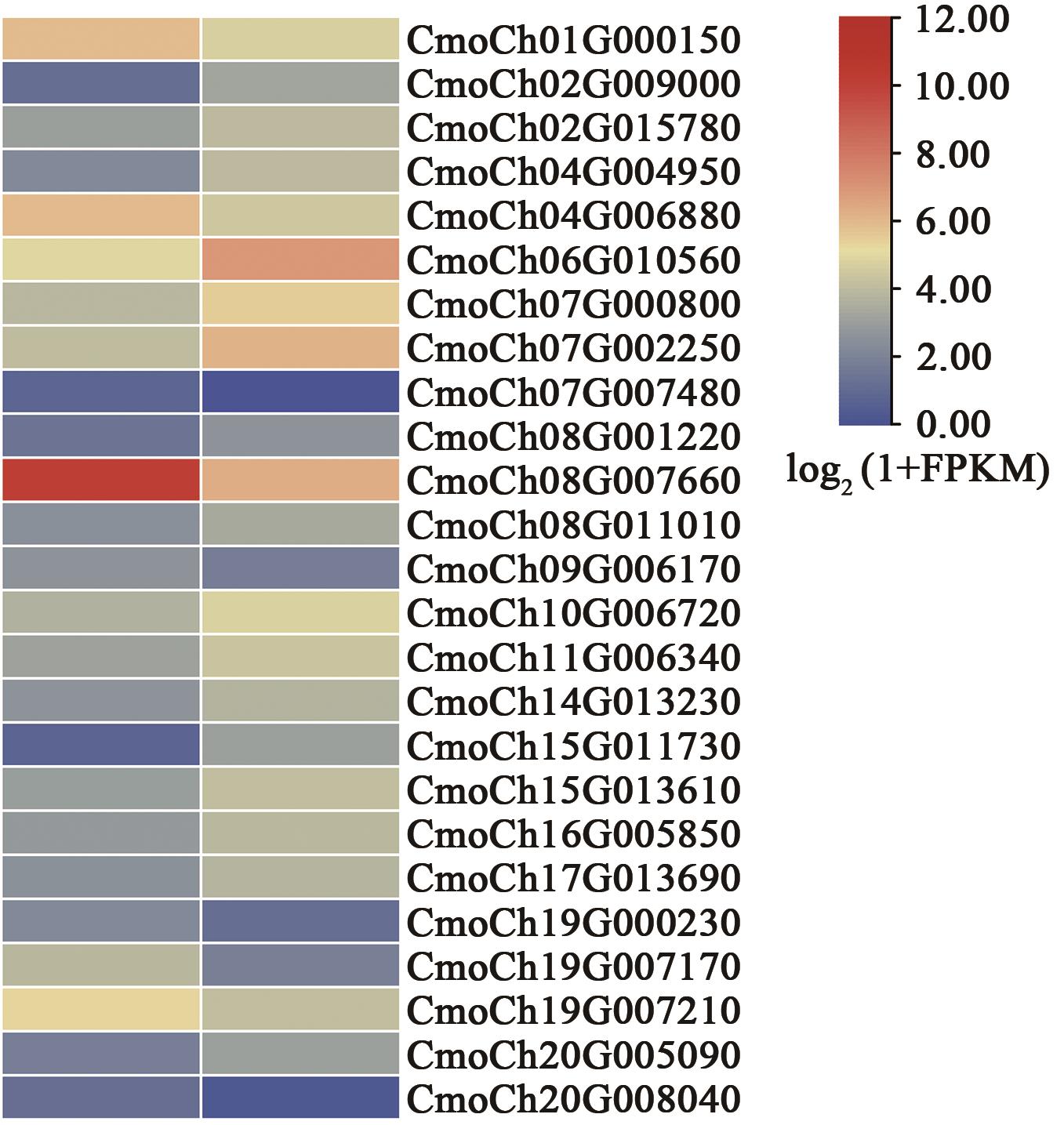
| 基因 ID | log2|FoldChange| | 基因起始位点 | 基因终止位点 | 注释 |
|---|---|---|---|---|
| CmoCh01G000150 | 1.192 986 569 | 51 481 | 52 456 | 生长素相关蛋白 |
| CmoCh02G009000 | -2.729 158 581 | 5 550 636 | 5 554 600 | 生长素应答因子22 |
| CmoCh02G015780 | -1.085 102 287 | 9 109 056 | 9 112 669 | 生长素渠化 |
| CmoCh04G004950 | -2.026 235 227 | 2 470 212 | 2 472 488 | 生长素响应GH3样蛋白1 |
| CmoCh04G006880 | 1.535 353 461 | 3 408 834 | 3 412 302 | 生长素诱导蛋白 |
| CmoCh06G010560 | -2.044 779 387 | 8 210 781 | 8 214 108 | 生长素响应GH3样蛋白5 |
| CmoCh07G000800 | -1.672 989 912 | 500 794 | 505 922 | 生长素应答因子 |
| CmoCh07G002250 | -2.181 053 727 | 1 136 983 | 1 140 904 | 生长素抑制蛋白 |
| CmoCh07G007480 | 3.270 328 289 | 3 388 548 | 3 390 222 | 生长素应答因子36 |
| CmoCh08G001220 | -1.678 882 862 | 629 552 | 633 268 | 生长素反应蛋白IAA26 |
| CmoCh08G007660 | 3.895 043 450 | 4 840 255 | 4 840 875 | 生长素结合蛋白 |
| CmoCh08G011010 | -1.027 143 495 | 7 056 498 | 7 063 138 | 生长素渠化 |
| CmoCh09G006170 | 1.054 262 364 | 3 084 770 | 3 091 533 | 生长素渠化 |
| CmoCh10G006720 | -1.210 110 125 | 3 087 573 | 3 089 725 | 生长素反应蛋白IAA27 |
| CmoCh11G006340 | -1.374 547 937 | 3 047 847 | 3 051 459 | 生长素反应蛋白IAA27 |
| CmoCh14G013230 | -1.248 495 558 | 11 047 643 | 11 049 773 | 生长素反应蛋白IAA26 |
| CmoCh15G011730 | -3.433 307 646 | 8 165 466 | 8 169 541 | 生长素渠化 |
| CmoCh15G013610 | -1.282 714 907 | 9 303 666 | 9 310 054 | 生长素应答因子9 |
| CmoCh16G005850 | -1.222 754 998 | 2 822 610 | 2 826 352 | 生长素应答因子9 |
| CmoCh17G013690 | -1.441 604 240 | 10 631 575 | 10 632 190 | 生长素反应蛋白 |
| CmoCh19G000230 | 1.584 356 252 | 139 153 | 141 383 | 生长素渠化 |
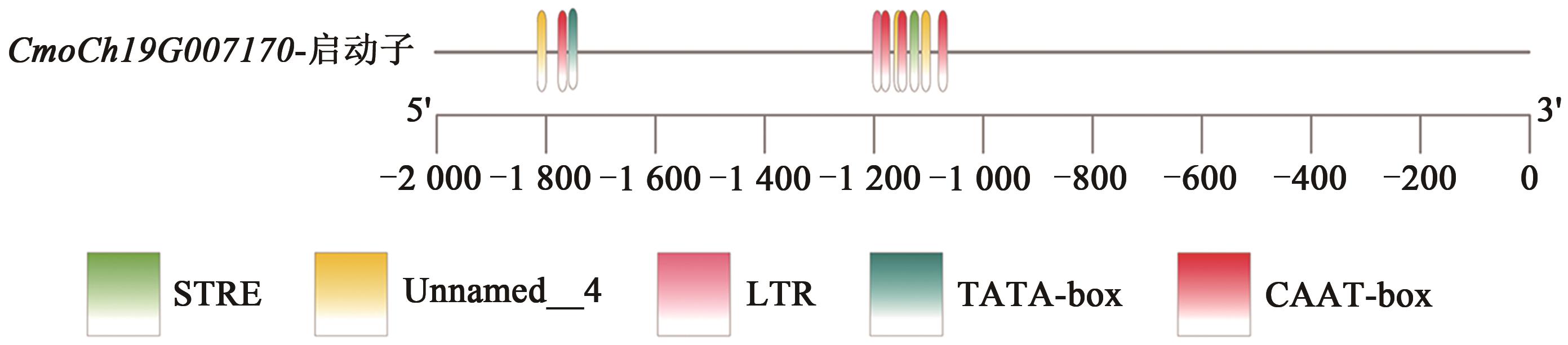
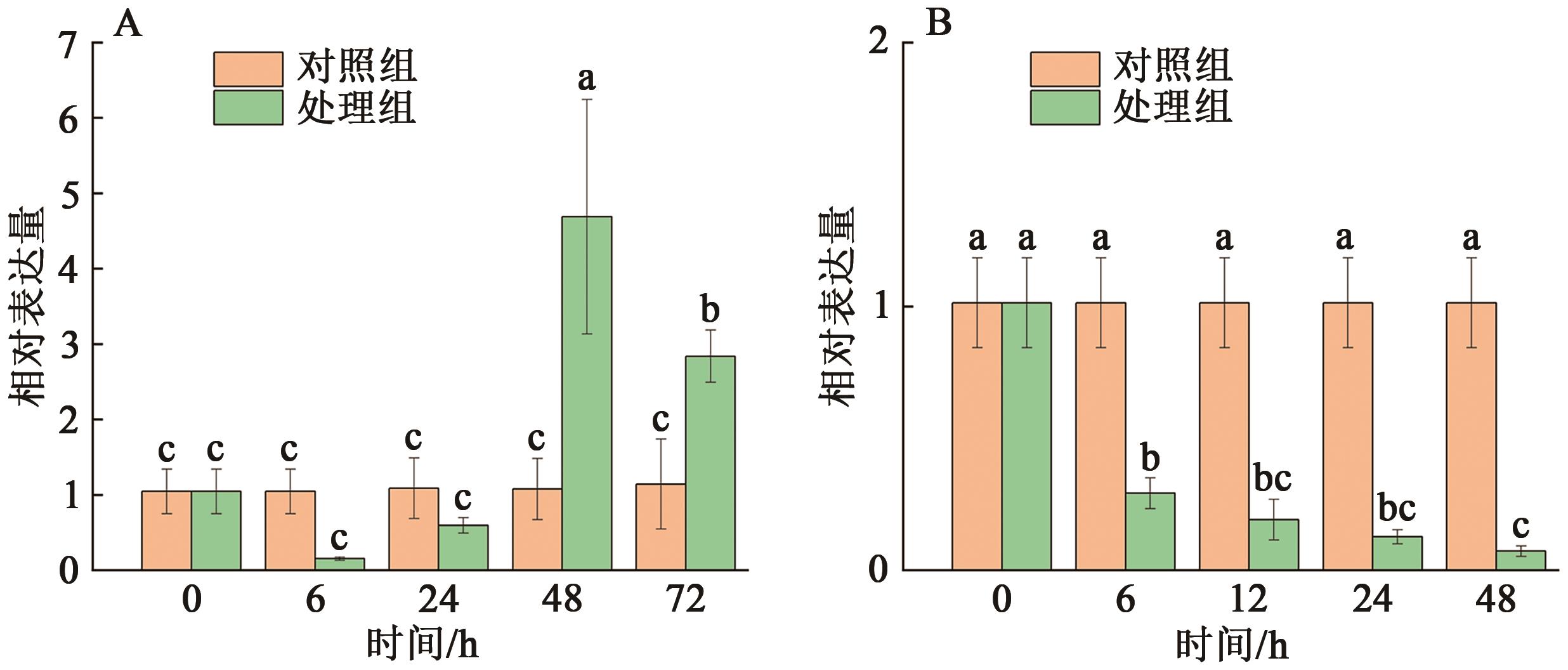
| CmoCh19G007170 | 2.359 833 611 | 7 501 456 | 7 501 755 | 生长素反应蛋白SAUR50 |
| CmoCh19G007210 | 1.222 892 554 | 7 514 737 | 7 515 027 | 生长素诱导蛋白 |
| CmoCh20G005090 | -1.540 231 132 | 2 433 984 | 2 440 115 | 生长素应答因子4 |
| CmoCh20G008040 | 2.795 275 878 | 3 960 038 | 3 962 188 | 生长素反应蛋白SAUR24 |
Table 1 Differential expression levels and annotation information of genes related to the IAA signaling pathway
| 基因 ID | log2|FoldChange| | 基因起始位点 | 基因终止位点 | 注释 |
|---|---|---|---|---|
| CmoCh01G000150 | 1.192 986 569 | 51 481 | 52 456 | 生长素相关蛋白 |
| CmoCh02G009000 | -2.729 158 581 | 5 550 636 | 5 554 600 | 生长素应答因子22 |
| CmoCh02G015780 | -1.085 102 287 | 9 109 056 | 9 112 669 | 生长素渠化 |
| CmoCh04G004950 | -2.026 235 227 | 2 470 212 | 2 472 488 | 生长素响应GH3样蛋白1 |
| CmoCh04G006880 | 1.535 353 461 | 3 408 834 | 3 412 302 | 生长素诱导蛋白 |
| CmoCh06G010560 | -2.044 779 387 | 8 210 781 | 8 214 108 | 生长素响应GH3样蛋白5 |
| CmoCh07G000800 | -1.672 989 912 | 500 794 | 505 922 | 生长素应答因子 |
| CmoCh07G002250 | -2.181 053 727 | 1 136 983 | 1 140 904 | 生长素抑制蛋白 |
| CmoCh07G007480 | 3.270 328 289 | 3 388 548 | 3 390 222 | 生长素应答因子36 |
| CmoCh08G001220 | -1.678 882 862 | 629 552 | 633 268 | 生长素反应蛋白IAA26 |
| CmoCh08G007660 | 3.895 043 450 | 4 840 255 | 4 840 875 | 生长素结合蛋白 |
| CmoCh08G011010 | -1.027 143 495 | 7 056 498 | 7 063 138 | 生长素渠化 |
| CmoCh09G006170 | 1.054 262 364 | 3 084 770 | 3 091 533 | 生长素渠化 |
| CmoCh10G006720 | -1.210 110 125 | 3 087 573 | 3 089 725 | 生长素反应蛋白IAA27 |
| CmoCh11G006340 | -1.374 547 937 | 3 047 847 | 3 051 459 | 生长素反应蛋白IAA27 |
| CmoCh14G013230 | -1.248 495 558 | 11 047 643 | 11 049 773 | 生长素反应蛋白IAA26 |
| CmoCh15G011730 | -3.433 307 646 | 8 165 466 | 8 169 541 | 生长素渠化 |
| CmoCh15G013610 | -1.282 714 907 | 9 303 666 | 9 310 054 | 生长素应答因子9 |
| CmoCh16G005850 | -1.222 754 998 | 2 822 610 | 2 826 352 | 生长素应答因子9 |
| CmoCh17G013690 | -1.441 604 240 | 10 631 575 | 10 632 190 | 生长素反应蛋白 |
| CmoCh19G000230 | 1.584 356 252 | 139 153 | 141 383 | 生长素渠化 |
| CmoCh19G007170 | 2.359 833 611 | 7 501 456 | 7 501 755 | 生长素反应蛋白SAUR50 |
| CmoCh19G007210 | 1.222 892 554 | 7 514 737 | 7 515 027 | 生长素诱导蛋白 |
| CmoCh20G005090 | -1.540 231 132 | 2 433 984 | 2 440 115 | 生长素应答因子4 |
| CmoCh20G008040 | 2.795 275 878 | 3 960 038 | 3 962 188 | 生长素反应蛋白SAUR24 |
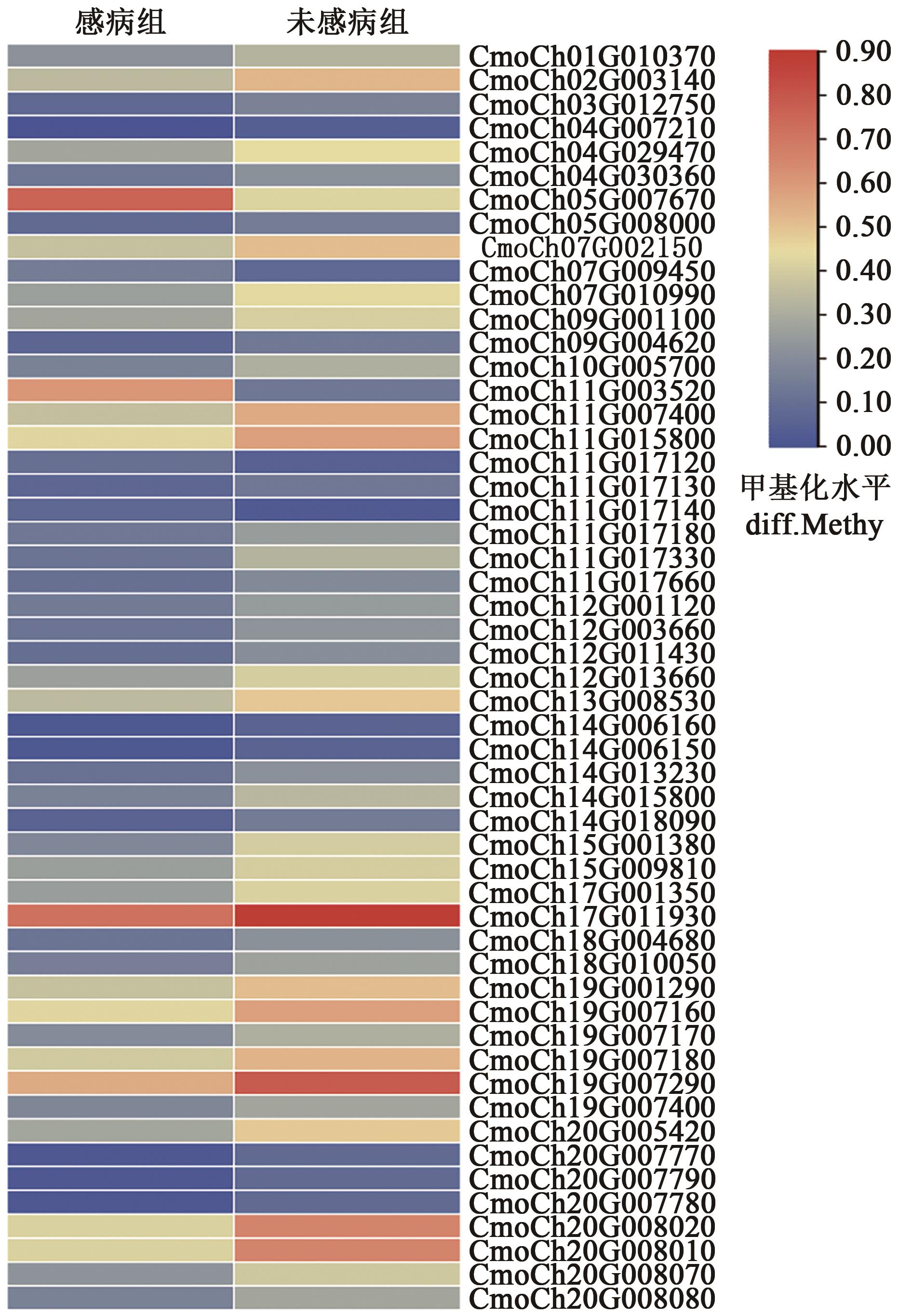
| 基因ID | 甲基化类型 | 甲基化区域 | 基因ID | 甲基化类型 | 甲基化区域 |
|---|---|---|---|---|---|
| CmoCh01G010370 | CHH | 内含子 | CmoCh13G008530 | CHH | 启动子 |
| CmoCh02G003140 | CHH | 内含子 | CmoCh14G006160 | CHH | 内含子 |
| CmoCh03G012750 | CHH | 启动子 | CmoCh14G006150 | CHH | 内含子 |
| CmoCh04G007210 | CHH | 启动子 | CmoCh14G013230 | CHH | 内含子 |
| CmoCh04G029470 | CHH | 启动子 | CmoCh14G015800 | CHH | 启动子 |
| CmoCh04G030360 | CHH | 启动子 | CmoCh14G018090 | CHH | 启动子 |
| CmoCh05G007670 | CHH, CG | 内含子, 外显子 | CmoCh15G001380 | CHH | 内含子 |
| CmoCh05G008000 | CHH | 启动子 | CmoCh15G009810 | CHH | 启动子 |
| CmoCh07G002150 | CHH | 启动子 | CmoCh17G001350 | CHH | 启动子 |
| CmoCh07G009450 | CHH | 启动子 | CmoCh17G011930 | CG, CHH | 启动子 |
| CmoCh07G010990 | CHH | 启动子 | CmoCh18G004680 | CHH | 内含子 |
| CmoCh09G001100 | CHH | 启动子 | CmoCh18G010050 | CHH | 内含子 |
| CmoCh09G004620 | CHH | 内含子 | CmoCh19G001290 | CHH | 启动子 |
| CmoCh10G005700 | CHH | 启动子 | CmoCh19G007160 | CHH | 启动子 |
| CmoCh11G003520 | CG | 外显子 | CmoCh19G007170 | CHH | 启动子 |
| CmoCh11G007400 | CG | 启动子 | CmoCh19G007180 | CHH | 启动子 |
| CmoCh11G015800 | CHH | 启动子 | CmoCh19G007290 | CG | 启动子 |
| CmoCh11G017120 | CHH | 启动子 | CmoCh19G007400 | CHH | 内含子, 启动子 |
| CmoCh11G017130 | CHH | 启动子 | CmoCh20G005420 | CHH | 启动子 |
| CmoCh11G017140 | CHH | 启动子 | CmoCh20G007770 | CHH | 启动子 |
| CmoCh11G017180 | CHH | 启动子 | CmoCh20G007790 | CHH | 启动子 |
| CmoCh11G017330 | CG, CHG | 内含子, 外显子 | CmoCh20G007780 | CHH | 内含子 |
| CmoCh11G017660 | CHH | 启动子 | CmoCh20G008020 | CG | 启动子 |
| CmoCh12G001120 | CHH | 内含子, 启动子 | CmoCh20G008010 | CG | 启动子 |
| CmoCh12G003660 | CHH | 启动子 | CmoCh20G008070 | CHH | 启动子 |
| CmoCh12G011430 | CHH | 启动子 | CmoCh20G008080 | CHH | 启动子 |
| CmoCh12G013660 | CHH | 启动子 |
Table 2 Methylation types and regions of genes to related IAA signaling pathway
| 基因ID | 甲基化类型 | 甲基化区域 | 基因ID | 甲基化类型 | 甲基化区域 |
|---|---|---|---|---|---|
| CmoCh01G010370 | CHH | 内含子 | CmoCh13G008530 | CHH | 启动子 |
| CmoCh02G003140 | CHH | 内含子 | CmoCh14G006160 | CHH | 内含子 |
| CmoCh03G012750 | CHH | 启动子 | CmoCh14G006150 | CHH | 内含子 |
| CmoCh04G007210 | CHH | 启动子 | CmoCh14G013230 | CHH | 内含子 |
| CmoCh04G029470 | CHH | 启动子 | CmoCh14G015800 | CHH | 启动子 |
| CmoCh04G030360 | CHH | 启动子 | CmoCh14G018090 | CHH | 启动子 |
| CmoCh05G007670 | CHH, CG | 内含子, 外显子 | CmoCh15G001380 | CHH | 内含子 |
| CmoCh05G008000 | CHH | 启动子 | CmoCh15G009810 | CHH | 启动子 |
| CmoCh07G002150 | CHH | 启动子 | CmoCh17G001350 | CHH | 启动子 |
| CmoCh07G009450 | CHH | 启动子 | CmoCh17G011930 | CG, CHH | 启动子 |
| CmoCh07G010990 | CHH | 启动子 | CmoCh18G004680 | CHH | 内含子 |
| CmoCh09G001100 | CHH | 启动子 | CmoCh18G010050 | CHH | 内含子 |
| CmoCh09G004620 | CHH | 内含子 | CmoCh19G001290 | CHH | 启动子 |
| CmoCh10G005700 | CHH | 启动子 | CmoCh19G007160 | CHH | 启动子 |
| CmoCh11G003520 | CG | 外显子 | CmoCh19G007170 | CHH | 启动子 |
| CmoCh11G007400 | CG | 启动子 | CmoCh19G007180 | CHH | 启动子 |
| CmoCh11G015800 | CHH | 启动子 | CmoCh19G007290 | CG | 启动子 |
| CmoCh11G017120 | CHH | 启动子 | CmoCh19G007400 | CHH | 内含子, 启动子 |
| CmoCh11G017130 | CHH | 启动子 | CmoCh20G005420 | CHH | 启动子 |
| CmoCh11G017140 | CHH | 启动子 | CmoCh20G007770 | CHH | 启动子 |
| CmoCh11G017180 | CHH | 启动子 | CmoCh20G007790 | CHH | 启动子 |
| CmoCh11G017330 | CG, CHG | 内含子, 外显子 | CmoCh20G007780 | CHH | 内含子 |
| CmoCh11G017660 | CHH | 启动子 | CmoCh20G008020 | CG | 启动子 |
| CmoCh12G001120 | CHH | 内含子, 启动子 | CmoCh20G008010 | CG | 启动子 |
| CmoCh12G003660 | CHH | 启动子 | CmoCh20G008070 | CHH | 启动子 |
| CmoCh12G011430 | CHH | 启动子 | CmoCh20G008080 | CHH | 启动子 |
| CmoCh12G013660 | CHH | 启动子 |
| 1 | PÉREZ-GARCÍA A, ROMERO D, FERNÁNDEZ-ORTUÑO D, et al.. The powdery mildew fungus Podosphaera fusca (synonym Podosphaera xanthii), a constant threat to cucurbits[J]. Mol. Plant Pathol., 2009, 10(2): 153-160. |
| 2 | BABADOOST M, SULLEY S, XIANG Y. Sensitivities of cucurbit powdery mildew fungus ( Podosphaera xanthii ) to fungicides[J]. Plant Hlth. Prog., 2020, 21:272-277. |
| 3 | BADRI A M, BAE I, LEE S. Marker-assisted evaluation of two powdery mildew resistance candidate genes in korean cucumber inbred lines[J]. Agronomy, 2021, 11(11):2191.?补 |
| 4 | 孙海,王晓青,李云龙,等.寡雄腐霉施用时期对设施黄瓜霜霉病的防治试验[J].生物技术进展,2014,4(1):63-65. |
| SUN H, WANG X Q, LI Y L, et al.. Evaluation of application time for Pythium oligandrum against cucumber downy mildew in greenhouse[J]. Curr. Biotechnol., 2014, 4(1): 63-65. | |
| 5 | 吴星波,郝俊杰,张晓艳,等.普通菜豆抗白粉病基因SCAR标记鉴定[J].生物技术进展,2013,3(5):357-362. |
| WU X B, HAO J J, ZHANG X Y, et al.. Identification of powdery mildew resistant genes based on SCAR markers in common bean (Phaseolus vulgaris L.)[J]. Curr. Biotechnol., 2013, 3(5): 357-362. | |
| 6 | 冯寒骞,李超.生长素信号转导研究进展[J].生物技术通报,2018,34(7):24-30. |
| FENG H Q, LI C. Research advances of auxin signal transduction[J]. Biotechnol. Bull., 2018, 34(7): 24-30. | |
| 7 | DHARMASIRI N, DHARMASIRI S, ESTELLE M. The F-box protein TIR1 is an auxin receptor[J]. Nature, 2005, 435: 441-445. |
| 8 | KEPINSKI S, LEYSER O. The Arabidopsis F-box protein TIR1 is an auxin receptor[J]. Nature, 2005, 435: 446-451. |
| 9 | TAN X, CALDERON-VILLALOBOS L I A, SHARON M, et al.. Mechanism of auxin perception by the TIR1 ubiquitin ligase[J]. Nature, 2007, 446: 640-645. |
| 10 | SALEHIN M, BAGCHI R, ESTELLE M. SCFTIR1/AFB-based auxin perception: mechanism and role in plant growth and development[J]. Plant Cell, 2015, 27(1): 9-19. |
| 11 | GRAY W M, KEPINSKI S, ROUSE D, et al.. Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins[J]. Nature, 2001, 414: 271-276. |
| 12 | CHEN M, NIE G, YANG L, et al.. Homeotic transformation from stamen to petal in Lilium is associated with MADS-box genes and hormone signal transduction[J]. Plant Growth Regul., 2021, 95(1): 49-64. |
| 13 | JIN L, QIN Q, WANG Y, et al.. Rice dwarf virus P2 protein hijacks auxin signaling by directly targeting the rice OsIAA10 protein, enhancing viral infection and disease development[J/OL]. PLoS Pathog., 2016, 12(9): e1005847[2024-03-15]. . |
| 14 | MEHMOOD A, HUSSAIN A, IRSHAD M, et al.. IAA and flavonoids modulates the association between maize roots and phytostimulant endophytic Aspergillus fumigatus greenish[J]. J. Plant Interact., 2018, 13(1):532-542. |
| 15 | HE Y, ZHANG T, SUN Y, et al.. Exogenous IAA alleviates arsenic toxicity to rice and reduces arsenic accumulation in rice grains[J]. J. Plant Growth. Regul., 2022, 41(2): 734-741. |
| 16 | WEN T, DONG L, WANG L, et al.. Changes in root architecture and endogenous hormone levels in two Malus rootstocks under alkali stress[J]. Sci. Hortic. Amsterdam., 2018, 235:198-204. |
| 17 | NAVARRO L, DUNOYER P, JAY F, et al.. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling[J]. Science, 2006, 312(5772): 436-439. |
| 18 | ZHANG Z, LI Q, LI Z, et al.. Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis-Pseudomonas syringae interaction[J]. Plant Physiol., 2007, 145(2): 450-464. |
| 19 | PADMANABHAN M S, SHIFERAW H, CULVER J N. The Tobacco mosaic virus replicase protein disrupts the localization and function of interacting Aux/IAA proteins[J]. Mol. Plant Microbe Interact., 2006, 19(8): 864-873. |
| 20 | SUN Y, FAN M, HE Y. DNA methylation analysis of the Citrullus lanatus response to Cucumber GreenMottle mosaic virus infection by whole-genome bisulfite sequencing[J/OL]. Genes Basel., 2019, 10(5): e344[2024-03-15]. . |
| 21 | RAMBANI A, RICE J H, LIU J, et al.. The methylome of soybean roots during the compatible interaction with the soybean cyst nematode[J]. Plant Physiol., 2015, 168(4): 1364-1377. |
| 22 | 徐妍.灰斑病菌胁迫对大豆生理生化和DNA甲基化的影响[D].哈尔滨:哈尔滨师范大学,2015. |
| 23 | CAO X, ZHAI X, ZHAO Z, et al.. Genome-wide DNA methylation analysis of paulownia with phytoplasma infection[J/OL]. Gene, 2020, 755: 144905[2024-03-15]. . |
| 24 | WANG S, YAN W, YANG X, et al.. Comparative methylome reveals regulatory roles of DNA methylation in melon resistance to Podosphaera xanthii [J/OL]. Plant Sci., 2021, 309: 110954[2024-03-15]. . |
| 25 | ZITTER T A. Compendium of Cucurbit Diseases[M]. US: Amer Phytopathological Society, 1996. |
| 26 | 方汉顺,谢永盾,曾伟伟,等.小麦矮秆突变体jm22d响应赤霉素处理的转录组学分析[J].生物技术进展,2020,10(5):503-516. |
| FANG H S, XIE Y D, ZENG W W, et al.. The transcriptome analysis of wheat dwarf mutant jm22d responding to GA treatment[J]. Curr. Biotechnol., 2020, 10(5): 503-516. | |
| 27 | MO H J, SUN Y X, ZHU X L, et al.. Cotton S-adenosylmethionine decarboxylase-mediated spermine biosynthesis is required for salicylic acid- and leucine-correlated signaling in the defense response to Verticillium dahliae [J]. Planta, 2016, 243(4): 1023-1039. |
| 28 | PARK S Y, FUNG P, NISHIMURA N, et al.. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins[J]. Science, 2009, 324(5930): 1068-1071. |
| 29 | WOODWARD A W, BARTEL B. Auxin: regulation, action, and interaction[J]. Ann. Bot., 2005, 95(5): 707-735. |
| 30 | 陈中惠,李景蕻.外源植物激素对高温胁迫下水培绿萝耐热性的影响[J].生物技术进展,2023,13(5):742-747. |
| CHEN Z H, LI J H. Effects of exogenous phytohormones on heat tolerance of Epipremnum aureum under hydroponic culture[J]. Curr. Biotechnol., 2023, 13(5): 742-747. | |
| 31 | LISCUM E, REED J W. Genetics of Aux/IAA and ARF action in plant growth and development[J]. Plant Mol. Biol., 2002, 49(3): 387-400. |
| 32 | HAGEN G, GUILFOYLE T. Auxin-responsive gene expression: genes, promoters and regulatory factors[J]. Plant Mol. Biol., 2002, 49(3-4): 373-385. |
| 33 | STASWICK P E, SERBAN B, ROWE M, et al.. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid [J]. Plant Cell, 2005, 17(2): 616-627. |
| 34 | NEWMAN T C, OHME-TAKAGI M, TAYLOR C B, et al.. DST sequences, highly conserved among plant SAUR genes, target reporter transcripts for rapid decay in tobacco[J]. Plant Cell, 1993, 5(6): 701-714. |
| 35 | LI Z G, CHEN H W, LI Q T, et al.. Three SAUR proteins SAUR 76, SAUR77 and SAUR78 promote plant growth in Arabidopsis[J/OL]. Sci. Rep., 2015, 5: 12477[2024-03-15]. . |
| 36 | DENG G, HUANG X, XIE L, et al.. Identification and expression of SAUR genes in the CAM plant Agave [J/OL]. Genes Basel., 2019, 10(7): e555[2024-03-15]. . |
| 37 | LIU Y, XIAO L, CHI J, et al.. Genome-wide identification and expression of SAUR gene family in peanut (Arachis hypogaea L.) and functional identification of AhSAUR3 in drought tolerance[J/OL]. BMC Plant Biol., 2022, 22(1): 178[2024-03-15]. . |
| [1] | AYELHAN·Haysa, Fei JIAO, Hong LIU, ANASI·Hudelati, Yubang SHEN. Integrating GWAS and Transcriptome Profiling to Identify SNP Markers Linked to High-temperature Tolerance in Esox lucius [J]. Current Biotechnology, 2025, 15(3): 432-445. |
| [2] | Zhuoying LIU, Xiaojin ZHOU, Yanli HUANG, Sen PANG. Joint Transcriptome Analysis of Maize Under Salt Stress and MeJA Treatment [J]. Current Biotechnology, 2025, 15(2): 263-275. |
| [3] | Yezi MA, Meijuan XIA, Cuicui LIU, Hongtao WANG, Jiaxi ZHOU. Comparative Analysis of Platelet Transcriptome and Proteome Changes in SARS-CoV2 Omicron Infection [J]. Current Biotechnology, 2024, 14(4): 649-656. |
| [4] | Hongmei QIAO. Transcriptome Analysis of the Terrestrial Plant Carallia brachiata from Rhizophoraceae [J]. Current Biotechnology, 2024, 14(2): 228-236. |
| [5] | Caihong WANG, Yuhan SHAO, Mengyuan JIANG. The Mechanisms of IL-17 Inhibiting Pseudomonas aeruginosa Infection on Lung Epithelial Cells [J]. Current Biotechnology, 2024, 14(2): 304-311. |
| [6] | Qinqin LIU, Baiming HUANG, Yezi MA, Cuicui LIU, Hongtao WANG, Jiaxi ZHOU. Comparative Analysis of Megakaryocyte and Platelet Transcriptome Changes in Acute Infection [J]. Current Biotechnology, 2023, 13(3): 465-472. |
| [7] | Dongao LI, Huiquan LIU, Qinhu WANG. Research Progress on Wheat Transcriptomes Responsive to Fusarium graminearum Infection [J]. Current Biotechnology, 2021, 11(5): 610-617. |
| [8] | FANG Hanshun1,2, XIE Yongdun2, ZENG Weiwei2, GUO Huijun2, XIONG Hongchun2, ZHAO Linshu2, GU Jiayu2, XU Yanhao1*, LIU Luxiang2*. The Transcriptome Analysis of Wheat Dwarf Mutant jm22d Responding to GA Treatment [J]. Curr. Biotech., 2020, 10(5): 503-516. |
| [9] | HUANG Chunmeng1,2, ZHU Pengyu1, WANG Zhi1, WANG Chenguang1, DU Zhixin3, WEI Shuang4, ZHANG Yongjiang1, FU Wei1*. Progress on RNAi-based Transgenic Plants [J]. Curr. Biotech., 2020, 10(1): 1-9. |
| [10] | GU Pei-yun1, LU Run-gang1, WANG Bu-yun2, SUN Hai2, YANG Jin-li1, JIAO Xue-xia1, LI Yun-long2* . Study on Control Efficiency of Electrolyzed Functional Water Against Hot Pepper Powdery Mildew [J]. Curr. Biotech., 2016, 6(1): 71-74. |
| [11] | WU Xing-bo1,2, HAO Jun-jie1*, ZHANG Xiao-yan1, WAN Shu-wei1, LI Hong-wei1, SHAO Yang1, SUN Ji-lu1. Identification of Powdery Mildew Resistant Genes based on SCAR Markers in Common Bean (Phaseolus vulgaris L.) [J]. Curr. Biotech., 2013, 3(5): 357-362. |
| [12] | QIN Tai-chen. A Summarized Opinion on Application of Crop Male Sterility in Plant Breeding [J]. journal1, 2011, 1(2): 84-89. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||