Current Biotechnology ›› 2024, Vol. 14 ›› Issue (2): 248-256.DOI: 10.19586/j.2095-2341.2024.0007
• Articles • Previous Articles Next Articles
The Sequence Characterization of the Somatic Footprints upon the Transposition of Non-autonomous Transposon rDt in Maize
Guantao ZHAO1,2( ), Jingsheng TAN1, Li ZHU1, Yubin LI1,3(
), Jingsheng TAN1, Li ZHU1, Yubin LI1,3( )
)
- 1.Biotechnology Research Institute,Chinese Academy of Agricultural Sciences,Beijing 100081,China
2.Gansu Yasheng Agricultural Research Institute Co. ,Ltd. ,Lanzhou 730030,China
3.College of Agronomy,Qingdao Agricultural University,Shandong Qingdao 266109,China
-
Received:2024-01-15Accepted:2024-02-07Online:2024-03-25Published:2024-04-17 -
Contact:Yubin LI
玉米非自主性转座子rDt的体细胞转座序列特征
- 1.中国农业科学院生物技术研究所,北京 100081
2.甘肃亚盛农业研究院有限公司,兰州 730030
3.青岛农业大学农学院,山东 青岛 266109
-
通讯作者:李玉斌 -
作者简介:赵官涛 E-mail: zhaoguantao@163.com; -
基金资助:国家自然科学基金面上项目(32072011);青岛农业大学高层次人才科研基金项目(665/1120002)
CLC Number:
Cite this article
Guantao ZHAO, Jingsheng TAN, Li ZHU, Yubin LI. The Sequence Characterization of the Somatic Footprints upon the Transposition of Non-autonomous Transposon rDt in Maize[J]. Current Biotechnology, 2024, 14(2): 248-256.
赵官涛, 谭景胜, 朱莉, 李玉斌. 玉米非自主性转座子rDt的体细胞转座序列特征[J]. 生物技术进展, 2024, 14(2): 248-256.
share this article
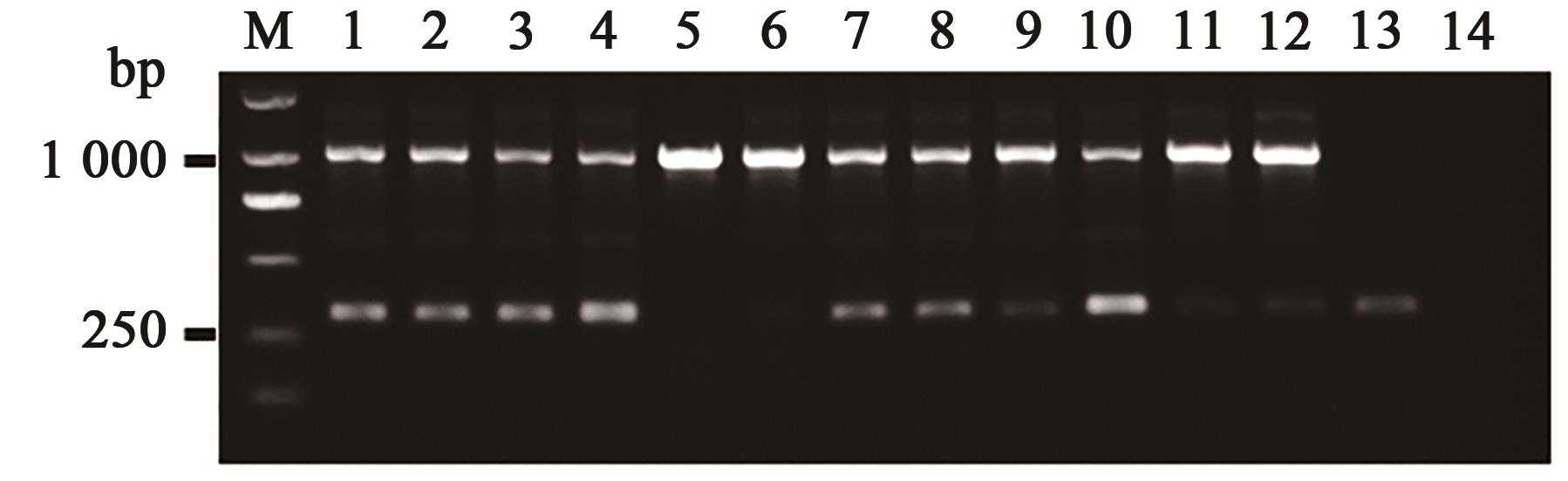
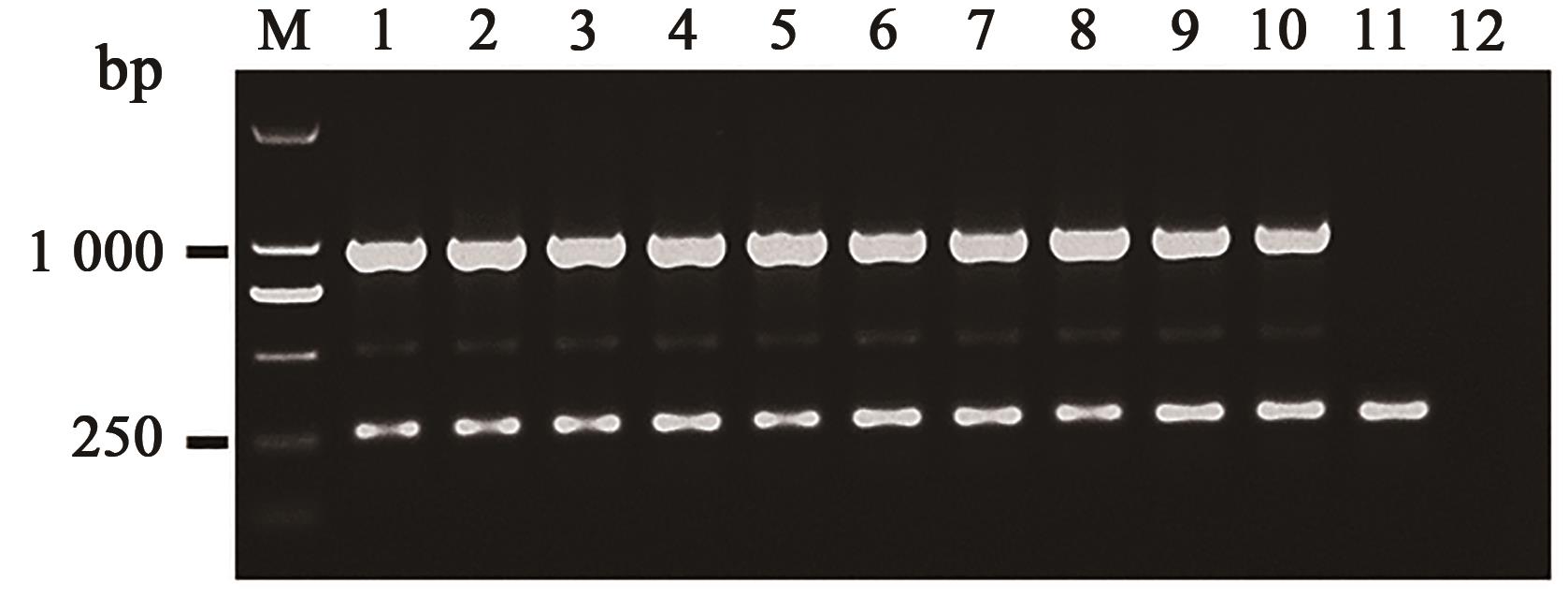
| 引物名称 | 引物序列(5'→3') |
|---|---|
| A1-1 | 5'-GCGAACGTTGGGAAGACGA-3' |
| A1-2 | 5'-CACCAGAGCCTCTACAGAT-3' |
| A1-3 | 5'-ATGAGCTGCACCTGCTTGAG-3' |
| A1-4 | 5'-TCAACTGAACTTCGACGACG-3' |
| A1-8 | 5'-ATGAGGTAATCAAGCCGACG-3' |
| A1-9 | 5'-GACACGAAGTACATCTGCAG-3' |
| A1-10 | 5'-GCTGCACAATTAGTCTCTCG-3' |
| A1-11 | 5'-TCAGTTGAATTGATGAGGCG-3' |
Table 1 Primers for PCR amplification
| 引物名称 | 引物序列(5'→3') |
|---|---|
| A1-1 | 5'-GCGAACGTTGGGAAGACGA-3' |
| A1-2 | 5'-CACCAGAGCCTCTACAGAT-3' |
| A1-3 | 5'-ATGAGCTGCACCTGCTTGAG-3' |
| A1-4 | 5'-TCAACTGAACTTCGACGACG-3' |
| A1-8 | 5'-ATGAGGTAATCAAGCCGACG-3' |
| A1-9 | 5'-GACACGAAGTACATCTGCAG-3' |
| A1-10 | 5'-GCTGCACAATTAGTCTCTCG-3' |
| A1-11 | 5'-TCAGTTGAATTGATGAGGCG-3' |
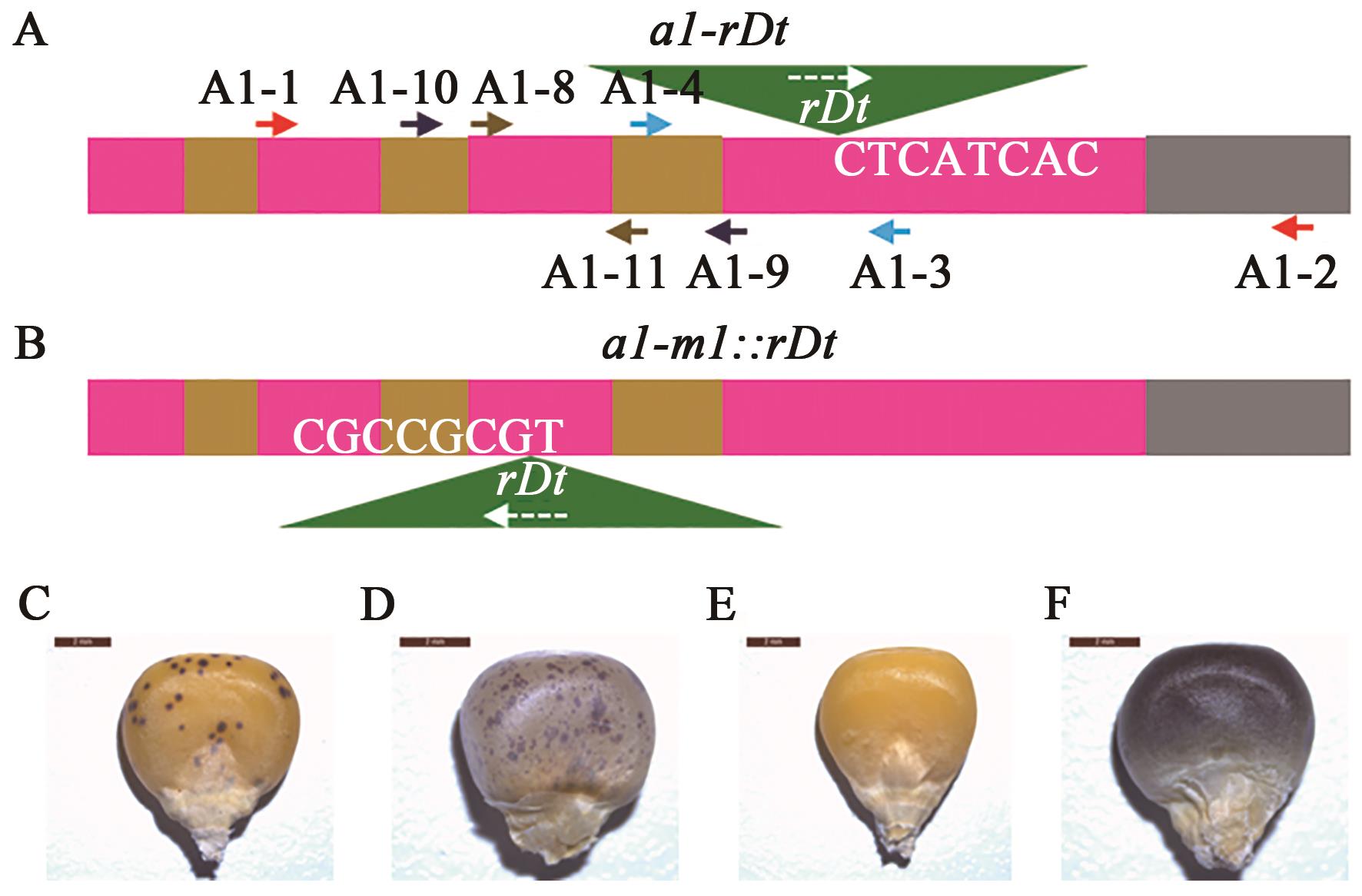
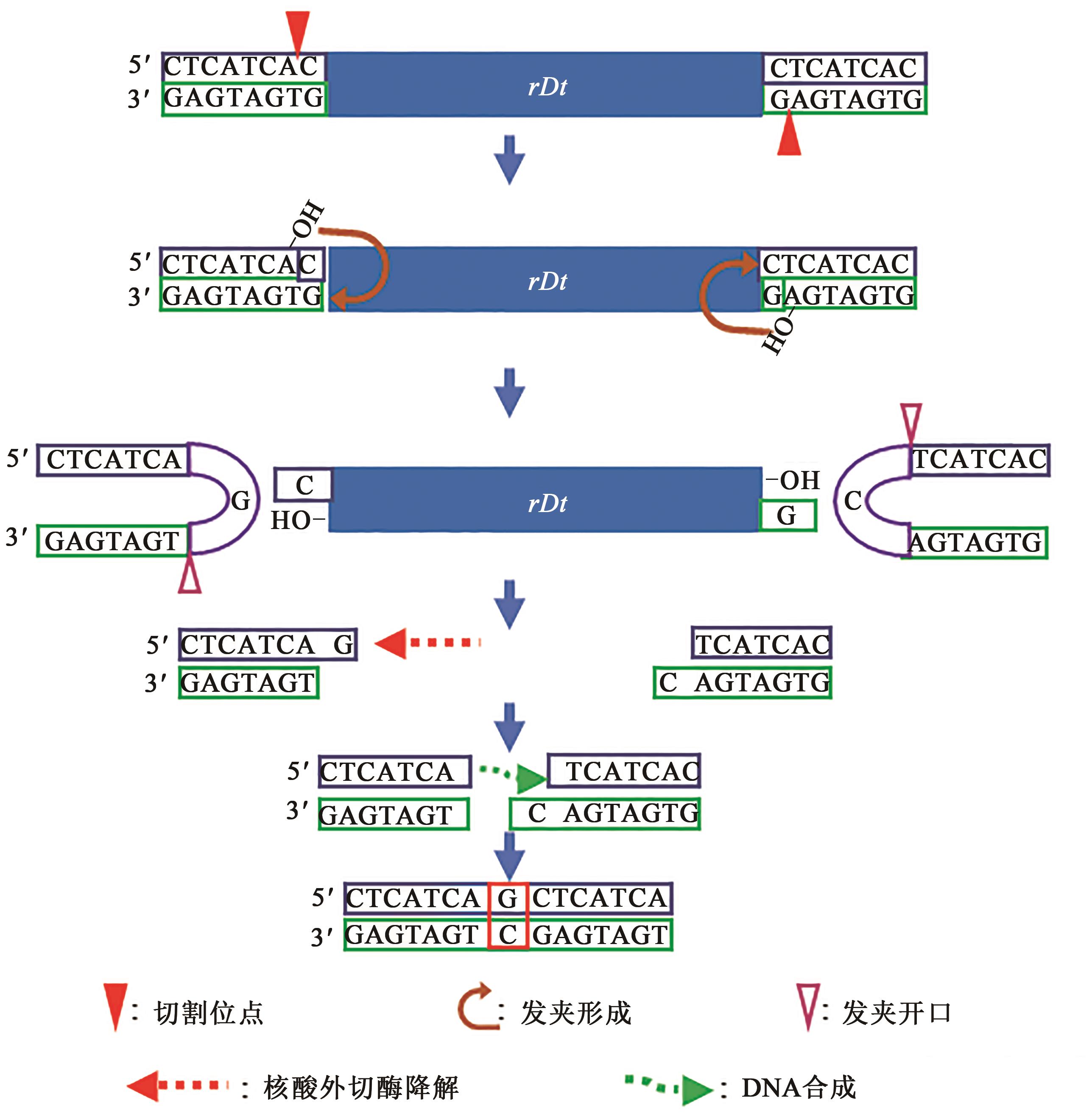
| 基因型 | 序列组成 | 变异类别 |
|---|---|---|
| A1 | 5′-GCCCAGC | 野生型 |
| a1-rDt | 5′-GCCCAGC | 插入突变 |
| a1-rDt-SE1(9) | 5′-GCCCAGC | 8+0 |
| a1-rDt-SE2(40) | 5′-GCCCAGC | 7+1+7(8+7) |
Table 2 Footprints analysis of a1-rDt somatic excision (SE)
| 基因型 | 序列组成 | 变异类别 |
|---|---|---|
| A1 | 5′-GCCCAGC | 野生型 |
| a1-rDt | 5′-GCCCAGC | 插入突变 |
| a1-rDt-SE1(9) | 5′-GCCCAGC | 8+0 |
| a1-rDt-SE2(40) | 5′-GCCCAGC | 7+1+7(8+7) |
| 基因型 | 序列组成 | 变异类别 |
|---|---|---|
| A1 | 5′-CACCGT | 野生型 |
| a1-m1::rDt | 5′-CACCGT | 插入突变 |
| a1-m1-SE1(20) | 5′-CACCGT | 7+7(8+6) |
| a1-m1-SE2(10) | 5′-CACCGT | 7+2+7(8+8) |
| a1-m1-SE3(1) | 5′-CACCGT | 7+3+7(8+9) |
| a1-m1-SE4(1) | 5′-CACCGT | 7+1+6(8+6) |
| a1-m1-SE5(1) | 5′-CACC............. | -2+5(8-5) |
Table 3 Footprints analysis of a1-m1::rDt somatic excision(SE)
| 基因型 | 序列组成 | 变异类别 |
|---|---|---|
| A1 | 5′-CACCGT | 野生型 |
| a1-m1::rDt | 5′-CACCGT | 插入突变 |
| a1-m1-SE1(20) | 5′-CACCGT | 7+7(8+6) |
| a1-m1-SE2(10) | 5′-CACCGT | 7+2+7(8+8) |
| a1-m1-SE3(1) | 5′-CACCGT | 7+3+7(8+9) |
| a1-m1-SE4(1) | 5′-CACCGT | 7+1+6(8+6) |
| a1-m1-SE5(1) | 5′-CACC............. | -2+5(8-5) |
| 杂交组合群体 | 群体粒数 | A1' | 回复突变频率 | A1'序列组成 |
|---|---|---|---|---|
| a1-rDt;Dt×a1-rMrh;+ | 135 240 | 16 | 1.19×10-4 | a1-rDt-SE1 |
| a1-rMrh;+×a1-rDt;Dt | 14 950 | 4 | 2.67×10-4 | a1-rDt-SE1 |
| a1-m1::rDt;Dt×a1-rMrh;+ | 26 100 | 14 | 5.39×10-4 | a1-m1-SE1 |
| a1-rMrh;+×a1-m1::rDt;Dt | 5 600 | 10 | 1.79×10-3 | a1-m1-SE1 |
Table 4 A1' reversion frequency analysis of different a1-m crosses and reciprocal crosses
| 杂交组合群体 | 群体粒数 | A1' | 回复突变频率 | A1'序列组成 |
|---|---|---|---|---|
| a1-rDt;Dt×a1-rMrh;+ | 135 240 | 16 | 1.19×10-4 | a1-rDt-SE1 |
| a1-rMrh;+×a1-rDt;Dt | 14 950 | 4 | 2.67×10-4 | a1-rDt-SE1 |
| a1-m1::rDt;Dt×a1-rMrh;+ | 26 100 | 14 | 5.39×10-4 | a1-m1-SE1 |
| a1-rMrh;+×a1-m1::rDt;Dt | 5 600 | 10 | 1.79×10-3 | a1-m1-SE1 |
| 1 | RAVINDRAN S. Barbara McClintock and the discovery of jumping genes[J]. Proc. Natl. Acad. Sci. USA, 2012, 109(50): 20198-20199. |
| 2 | MCCLINTOCK B. The origin and behavior of mutable loci in maize[J]. Proc. Natl. Acad. Sci. USA, 1950, 36(6): 344-355. |
| 3 | MOSCHETTI R, PALAZZO A, LORUSSO P, et al.. "What you need, baby, I got it": transposable elements as suppliers of cis-operating sequences in Drosophila [J/OL]. Biology, 2020, 9(2): 25[2023-12-06]. . |
| 4 | HAN M, PERKINS M H, NOVAES L S, et al.. Advances in transposable elements: from mechanisms to applications in mammalian genomics[J/OL]. Front. Genet., 2023, 14: 1290146[2023-12-06]. . |
| 5 | HAYWARD A, GILBERT C. Transposable elements[J]. Curr. Biol., 2022, 32(17): 904-909. |
| 6 | HUA-VAN A, LE ROUZIC A, BOUTIN T S, et al.. The struggle for life of the genome's selfish architects[J/OL]. Biol. Direct., 2011, 6(1): 19[2023-12-06]. . |
| 7 | GASPAROTTO E, BURATTIN F V, DI GIOIA V, et al.. Transposable elements co-option in genome evolution and gene regulation[J/OL]. Int. J. Mol. Sci., 2023, 24(3): 2610[2023-12-06]. . |
| 8 | COLONNA ROMANO N, FANTI L. Transposable elements: major players in shaping genomic and evolutionary patterns[J/OL]. Cells, 2022, 11(6): 1048[2023-12-06]. . |
| 9 | SAHEBI M, HANAFI M M, VAN WIJNEN A J, et al.. Contribution of transposable elements in the plant's genome[J]. Gene, 2018, 665: 155-166. |
| 10 | WICKER T, SABOT F, HUA-VAN A, et al.. A unified classification system for eukaryotic transposable elements[J]. Nat. Rev. Genet., 2007, 8(12): 973-982. |
| 11 | KAPITONOV V V, JURKA J. A universal classification of eukaryotic transposable elements implemented in Repbase[J]. Nat. Rev. Genet., 2008, 9(5): 411-412, 414. |
| 12 | MEISSNER R, CHAGUE V, ZHU Q, et al.. Technical advance: a high throughput system for transposon tagging and promoter trapping in tomato[J]. Plant J., 2000, 22(3): 265-274. |
| 13 | CARTER J D, PEREIRA A, DICKERMAN A W, et al.. An active ac/ds transposon system for activation tagging in tomato cultivar m82 using clonal propagation[J]. Plant Physiol., 2013, 162(1): 145-156. |
| 14 | LAZAROW K, DOLL M L, KUNZE R. Molecular biology of maize Ac/Ds elements: an overview[J]. Methods Mol. Biol., 2013, 1057: 59-82. |
| 15 | LI Y B, SEGAL G, WANG Q H, et al.. Gene tagging with engineered Ds elements in maize[J]. Methods Mol. Biol., 2013, 1057: 83-99. |
| 16 | WU H, XUE X, QIN C, et al.. An efficient system for Ds transposon tagging in Brachypodium distachyon [J]. Plant Physiol., 2019, 180(1): 56-65. |
| 17 | PETERSON T. Maize transposon storm kicks up a White cap [J]. Genetics, 2017, 206(1): 87-89. |
| 18 | LISCH D. Mutator and MULE transposons[J/OL]. Microbiol. Spectr., 2015, 3(2): MDN A3-0032-2014[2023-12-06]. . |
| 19 | GRABUNDZIJA I, MESSING S A, THOMAS J, et al.. A Helitron transposon reconstructed from bats reveals a novel mechanism of genome shuffling in eukaryotes[J/OL]. Nat. Commun., 2016, 7(1): 10716[2023-12-06]. . |
| 20 | KUNZE R, WEIL C F. The hAT and CACTA superfamilies of plant transposons[M]// CRAIG N L, CRAIGIE R, GELLERT M, et al.. Mobile DNA II. Washington, DC:ASM Press, 2002: 565-610. |
| 21 | RHOADES M M. The effect of varying gene dosage on aleurone colour in maize[J]. J. Genet., 1936, 33(3): 347-354. |
| 22 | BROWN J J, MATTES M G, O'REILLY C, et al.. Molecular characterization of rDt, a maize transposon of the "Dotted" controlling element system[J]. Mol. Gen. Genet., 1989, 215(2): 239-244. |
| 23 | CONG C S, LI Y B. Progress on mutator superfamily[J]. Hereditas, 2020, 42(2): 131-144. |
| 24 | ZHOU L, MITRA R, ATKINSON P W, et al.. Transposition of hAT elements links transposable elements and V(D)J recombination[J]. Nature, 2004, 432(7020): 995-1001. |
| 25 | XU Z, DOONER H K. Mx-rMx, a family of interacting transposons in the growing hAT superfamily of maize[J]. Plant Cell, 2005, 17(2): 375-388. |
| 26 | AYLON Y, KUPIEC M. DSB repair: the yeast paradigm[J]. DNA Repair, 2004, 3(8-9): 797-815. |
| 27 | RHOADES M M. Effect of the Dt gene on the mutability of the a(1) allele in maize[J]. Genetics, 1938, 23(4): 377-397. |
| 28 | RHOADES M M. On the genetic control of mutability in maize[J]. Proc. Natl. Acad. Sci. USA, 1945, 31(3): 91-95. |
| 29 | NAVARRO C. The mobile world of transposable elements[J]. Trends Genet., 2017, 33(11): 771-772. |
| 30 | MUÑOZ-LÓPEZ M, GARCÍA-PÉREZ J L. DNA transposons: nature and applications in genomics[J]. Curr. Genom. 2010, 11(2): 115-128. |
| 31 | GUFFANTI G, BARTLETT A, DECRESCENZO P, et al.. Transposable elements[J]. Curr. Top. Behav. Neurosci., 2019, 42: 221-246. |
| 32 | MA Y, PANNICKE U, SCHWARZ K, et al.. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination[J]. Cell, 2002, 108(6): 781-794. |
| 33 | NUFFER M G. Mutation studies at the A. locus in maize. I. A mutable allele controlled by Dt[J]. Genetics, 1961, 46(6): 625-640. |
| 34 | BRADLEY D, CARPENTER R, SOMMER H, et al.. Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of Antirrhinum [J]. Cell, 1993, 72(1): 85-95. |
| 35 | RINEHART T A, DEAN C, WEIL C F. Comparative analysis of non-random DNA repair following Ac transposon excision in maize and Arabidopsis [J]. Plant J., 1997, 12(6): 1419-1427. |
| [1] | Zhuoying LIU, Xiaojin ZHOU, Yanli HUANG, Sen PANG. Joint Transcriptome Analysis of Maize Under Salt Stress and MeJA Treatment [J]. Current Biotechnology, 2025, 15(2): 263-275. |
| [2] | Qingyun ZHANG, Lei MA, Hua XU, Lian JIN, Junjie ZOU, Baobao WANG, Quanjia CHEN, Miaoyun XU. Mechanism of Lignin Content in Root System Affecting Salt Tolerance in Maize [J]. Current Biotechnology, 2025, 15(1): 67-77. |
| [3] | Xin QI, Xinran LI, Yaning GUO, Dan WANG, Kai LI, Qiong WU, Liang LI. Comparison of Endogenous Genes in Maize Based on Digital PCR [J]. Current Biotechnology, 2025, 15(1): 78-85. |
| [4] | Jingsheng TAN, Guantao ZHAO, Li ZHU, Yubin LI. The Sequence Characterization of Somatic Excision Footprints from Two-element Transposable System of rMrh/Mrh in Maize [J]. Current Biotechnology, 2024, 14(4): 601-609. |
| [5] | Lingyan LI, Bing XIAO, Xudong ZHANG, Hua ZHANG, Ziyan CHEN, Haoqian WANG, Xiujie ZHANG, Hong CHEN, Jingang LIANG. Establishment and Standardization of Real-time PCR Method for Qualitative Detection of Genetically Modified Maize MON87411 [J]. Current Biotechnology, 2024, 14(2): 257-262. |
| [6] | Min LI, Lei WANG, Junjie ZOU. Opportunities and Challenges for the Industrial Application of Transgenic Insect-resistant and Herbicide-tolerant Maize in China [J]. Current Biotechnology, 2023, 13(2): 157-165. |
| [7] | Ke XIAO, Xiaojin ZHOU, Rumei CHEN, Sen PANG. Interactions Between ClassⅠand ClassⅡ NAS Proteins in Maize Using Bimolecular Fluorescence Complementation (BiFC) Assay [J]. Current Biotechnology, 2022, 12(5): 728-736. |
| [8] | Xing DANG, Binwei ZHI, Kehao CAO, Tingting LIU, Biao CHEN, Yuanjie DING. Patent Analysis on Genetically Modified Maize Biological Breeding Technology and Development Suggestions [J]. Current Biotechnology, 2022, 12(4): 614-622. |
| [9] | Hongtao WEN, Yang YANG, Yijia DING, Ran YUAN, Ruiying ZHANG, Zhiyuan XU, Jingang LIANG. Establishment of Qualitative PCR Detection of Transgenic Insect⁃resistant Maize CM8101 [J]. Current Biotechnology, 2022, 12(2): 248-255. |
| [10] | Yuhan YUAN, Jiali FAN, Wenzhu YANG, Rumei CHEN. Photosynthetic Characteristics Studies of Three Maize Yellow Leaf Mutants [J]. Current Biotechnology, 2022, 12(1): 75-82. |
| [11] | Shulei LI, Miaoyun XU, Hongyan ZHENG, Lei WANG. Establishment and Application of CRISPR/Cpf1‑mediated Base Editing System [J]. Current Biotechnology, 2021, 11(6): 732-740. |
| [12] | REN Wen1, YANG Haixia2, CHEN Lizhu2, LI Yufeng2, LIU Ya1*. Establishment and Application of Nucleic Acid Chromatography for Rapid Detection of Transgenic Plants [J]. Curr. Biotech., 2020, 10(6): 680-687. |
| [13] | WEN Hongtao1§,YANG Yang1§,DING Yijia1,YUAN Ran1,ZHANG Xiujie2,ZHANG Ruiying1*. Qualitative PCR Methods for the Detection of Transgenic Herbicide-tolerant Maize G1105E-823C [J]. Curr. Biotech., 2020, 10(6): 688-695. |
| [14] | WU Wenyan§, LIU Xinxiang§, ZHOU Miaoyi*, LIU Jinxiang, LIU Ya. Establishment of Event-specific PCR Detection Method of Transgenic Maize Line 2A-5 [J]. Curr. Biotech., 2020, 10(4): 363-370. |
| [15] | WANG Yali1,2, DENG Dexiang1,2, WANG Yijun1,2*. Recent Advances of G-protein Research in Maize [J]. Curr. Biotech., 2019, 9(1): 1-5. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||