Current Biotechnology ›› 2025, Vol. 15 ›› Issue (4): 645-654.DOI: 10.19586/j.2095-2341.2025.0017
• Reviews • Previous Articles Next Articles
Research Progress in Spatial Transcriptomics Technology for Liver Disease Research
Lingfei WAN( ), Wenting PAN, Yuting YONG, Yuanshuai LI, yue ZHAO, Xinlong YAN(
), Wenting PAN, Yuting YONG, Yuanshuai LI, yue ZHAO, Xinlong YAN( )
)
- College of Chemistry and Life Science,Beijing University of Technology,Beijing 100124,China
-
Received:2025-02-14Accepted:2025-03-31Online:2025-07-25Published:2025-09-08 -
Contact:Xinlong YAN
空间转录组学在肝病研究中的应用进展
万令飞( ), 潘文婷, 雍雨婷, 李元帅, 赵悦, 阎新龙(
), 潘文婷, 雍雨婷, 李元帅, 赵悦, 阎新龙( )
)
- 北京工业大学化学与生命科学学院,北京 100124
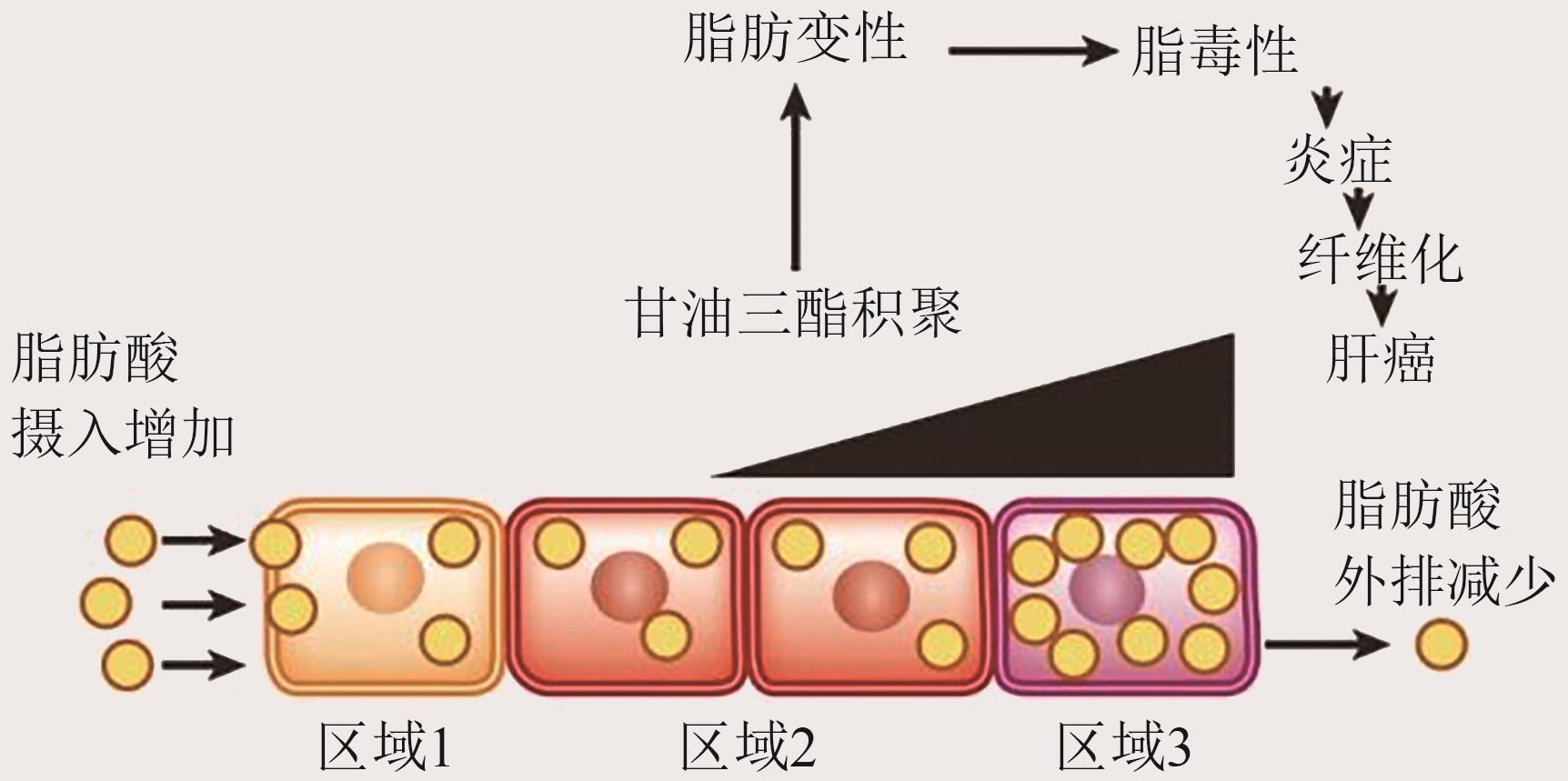
-
通讯作者:阎新龙 -
作者简介:万令飞 E-mail: wan-lingfei@outlook.com; -
基金资助:北京市自然科学基金和市教委基金联合基金重点项目(KZ202210005010)
CLC Number:
Cite this article
Lingfei WAN, Wenting PAN, Yuting YONG, Yuanshuai LI, yue ZHAO, Xinlong YAN. Research Progress in Spatial Transcriptomics Technology for Liver Disease Research[J]. Current Biotechnology, 2025, 15(4): 645-654.
万令飞, 潘文婷, 雍雨婷, 李元帅, 赵悦, 阎新龙. 空间转录组学在肝病研究中的应用进展[J]. 生物技术进展, 2025, 15(4): 645-654.
share this article
| 技术平台 | 分类 | 分辨率 | 检测区域 | 适用场景 | 优势 | 局限性 |
|---|---|---|---|---|---|---|
| 10×Genomics Visium | 测序型 | ~55 µm | 6.5×6.5 mm² | 组织大范围基因表达分析,适用于肝脏、肿瘤、神经组织 | 成熟商业化平台,实验流程标准化,支持FFPE样本 | 分辨率较低,每个点包含多个细胞,难以解析单细胞信息 |
| Slide-seq[ | 测序型 | ~10 µm | 6.2×6.2 mm² | 适用于神经系统、肝小叶分区研究 | 分辨率高于Visium,支持全转录组测序 | 捕获面积受限,微珠随机分布可能影响数据一致性 |
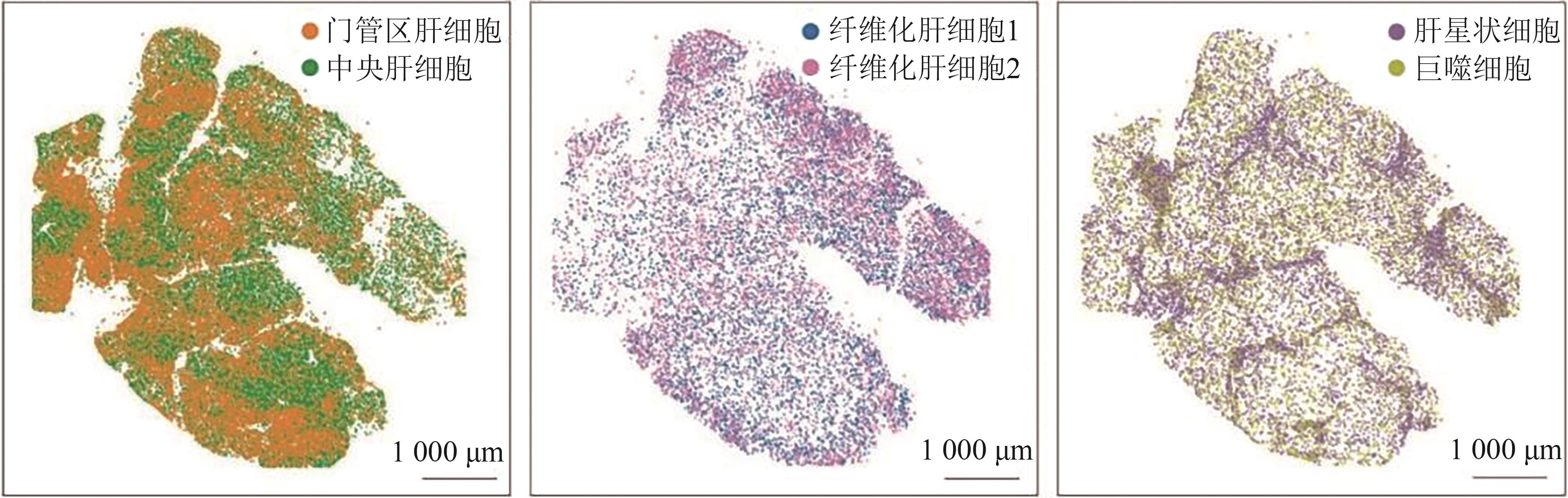
| Stereo-seq | 测序型 | ~500 nm | 最大可达13.2 cm² | 适用于超高分辨率组织解析,如胚胎发育、肿瘤 | 分辨率极高,可进行亚细胞级别分析,适用于大组织 | 数据处理复杂,计算量大,对实验条件要求高 |
| MERFISH | 成像型 | ~100 nm | 1~2 mm² | 适用于单细胞/亚细胞级别的基因表达研究,如神经科学、胚胎发育 | 超高分辨率,可检测10 000+目标基因 | 仅能检测预设基因,实验流程复杂,数据分析要求高 |
| SeqFISH | 成像型 | ~100 nm | 1~2 mm² | 适用于亚细胞级别的RNA成像,如免疫组织分析 | 高通量,可检测10 000+目标基因 | 仅适用于已知基因组,无法进行全转录组分析 |
| STARmap[ | 成像型 | ~200 nm | 1~3 mm² | 适用于单细胞分辨率的组织分析,如大脑结构 | 结合水凝胶扩增,提高信噪比,适合深层组织 | 基因检测数量有限,实验复杂度高 |
Table 1 Current mainstream spatial transcriptomics techniques
| 技术平台 | 分类 | 分辨率 | 检测区域 | 适用场景 | 优势 | 局限性 |
|---|---|---|---|---|---|---|
| 10×Genomics Visium | 测序型 | ~55 µm | 6.5×6.5 mm² | 组织大范围基因表达分析,适用于肝脏、肿瘤、神经组织 | 成熟商业化平台,实验流程标准化,支持FFPE样本 | 分辨率较低,每个点包含多个细胞,难以解析单细胞信息 |
| Slide-seq[ | 测序型 | ~10 µm | 6.2×6.2 mm² | 适用于神经系统、肝小叶分区研究 | 分辨率高于Visium,支持全转录组测序 | 捕获面积受限,微珠随机分布可能影响数据一致性 |
| Stereo-seq | 测序型 | ~500 nm | 最大可达13.2 cm² | 适用于超高分辨率组织解析,如胚胎发育、肿瘤 | 分辨率极高,可进行亚细胞级别分析,适用于大组织 | 数据处理复杂,计算量大,对实验条件要求高 |
| MERFISH | 成像型 | ~100 nm | 1~2 mm² | 适用于单细胞/亚细胞级别的基因表达研究,如神经科学、胚胎发育 | 超高分辨率,可检测10 000+目标基因 | 仅能检测预设基因,实验流程复杂,数据分析要求高 |
| SeqFISH | 成像型 | ~100 nm | 1~2 mm² | 适用于亚细胞级别的RNA成像,如免疫组织分析 | 高通量,可检测10 000+目标基因 | 仅适用于已知基因组,无法进行全转录组分析 |
| STARmap[ | 成像型 | ~200 nm | 1~3 mm² | 适用于单细胞分辨率的组织分析,如大脑结构 | 结合水凝胶扩增,提高信噪比,适合深层组织 | 基因检测数量有限,实验复杂度高 |
| [1] | MAYNARD K R, COLLADO-TORRES L, WEBER L M, et al.. Transcriptome-scale spatial gene expression in the human dorsolateral prefrontal cortex[J]. Nat. Neurosci., 2021, 24(3): 425-436. |
| [2] | CHEN A, LIAO S, CHENG M, et al.. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays[J]. Cell, 2022, 185(10): 1777-1792. |
| [3] | CHEN K H, BOETTIGER A N, MOFFITT J R, et al.. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells[J/OL]. Science, 2015, 348(6233): aaa6090[2025-06-24]. . |
| [4] | ENG C L, LAWSON M, ZHU Q, et al.. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH[J]. Nature, 2019, 568(7751): 235-239. |
| [5] | RODRIQUES S G, STICKELS R R, GOEVA A, et al.. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution[J]. Science, 2019, 363(6434): 1463-1467. |
| [6] | WANG X, ALLEN W E, WRIGHT M A, et al.. Three-dimensional intact-tissue sequencing of single-cell transcriptional states[J/OL]. Science, 2018, 361(6400): eaat5691[2025-06-24]. . |
| [7] | BERGMANN S, PENFOLD C A, SLATERY E, et al.. Spatial profiling of early primate gastrulation in utero [J]. Nature, 2022, 609(7925): 136-143. |
| [8] | MO C K, LIU J, CHEN S, et al.. Tumour evolution and microenvironment interactions in 2D and 3D space[J]. Nature, 2024, 634(8036): 1178-1186. |
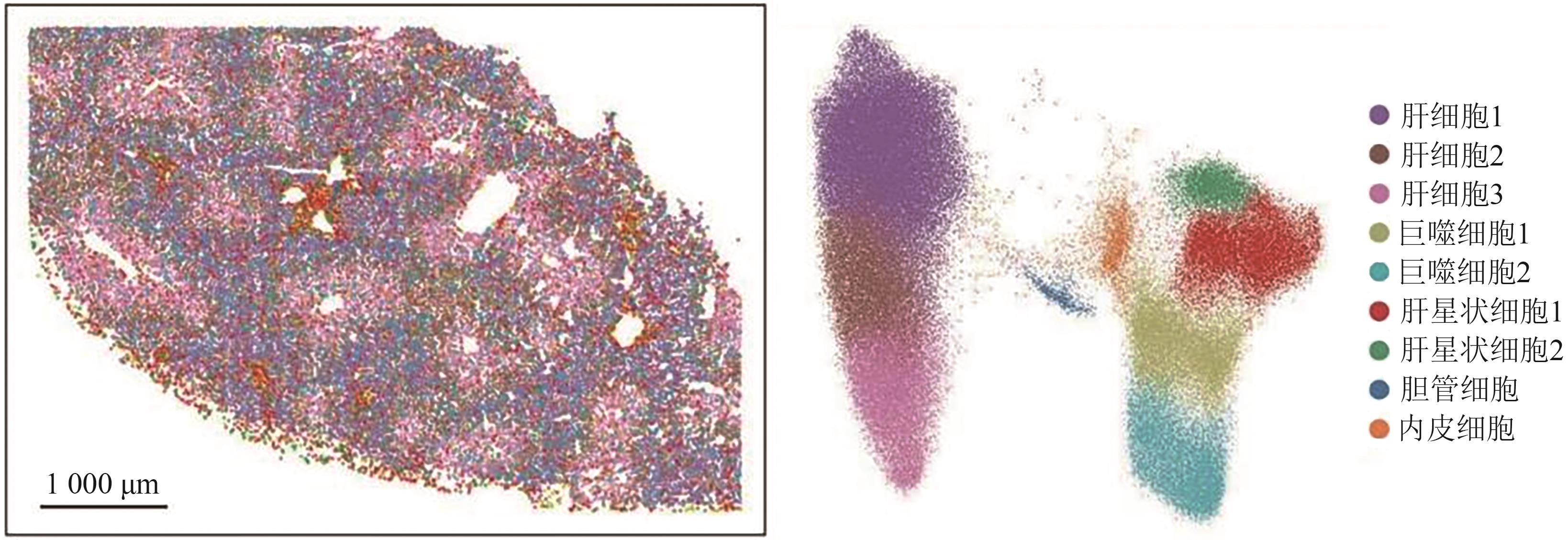
| [9] | DROIN C, KHOLTEI J E, BAHAR HALPERN K, et al.. Space-time logic of liver gene expression at sub-lobular scale[J]. Nat. Metab., 2021, 3(1): 43-58. |
| [10] | HALPERN K B, SHENHAV R, MATCOVITCH-NATAN O, et al.. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver[J]. Nature, 2017, 542(7641): 352-356. |
| [11] | GEBHARDT R, MATZ-SOJA M. Liver zonation: novel aspects of its regulation and its impact on homeostasis[J]. World J. Gastroenterol., 2014, 20(26): 8491-8504. |
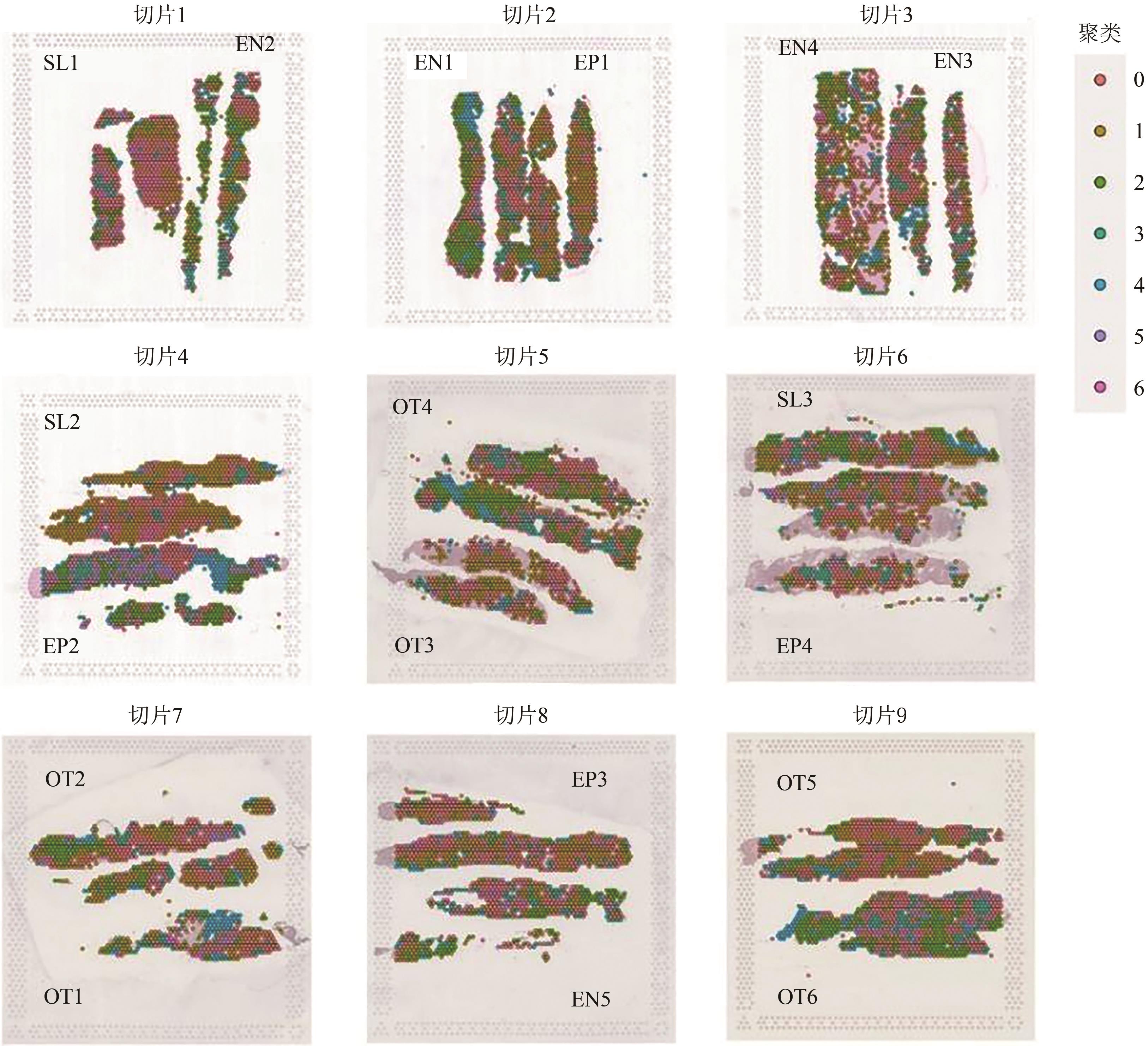
| [12] | WATSON B R, PAUL B, RAHMAN R U, et al.. Spatial transcriptomics of healthy and fibrotic human liver at single-cell resolution[J/OL]. Nat. Commun., 2025, 16(1): 319[2025-06-24]. . |
| [13] | RIAZI K, AZHARI H, CHARETTE J H, et al.. The prevalence and incidence of NAFLD worldwide: a systematic review and meta-analysis[J]. Lancet Gastroenterol. Hepatol., 2022, 7(9): 851-861. |
| [14] | SAVIANO A, HENDERSON N C, BAUMERT T F. Single-cell genomics and spatial transcriptomics: discovery of novel cell states and cellular interactions in liver physiology and disease biology[J]. J. Hepatol., 2020, 73(5): 1219-1230. |
| [15] | MARTINI T, NAEF F, TCHORZ J S. Spatiotemporal metabolic liver zonation and consequences on pathophysiology[J]. Annu. Rev. Pathol., 2023, 18: 439-466. |
| [16] | HUBY T, GAUTIER E L. Immune cell-mediated features of non-alcoholic steatohepatitis[J]. Nat. Rev. Immunol., 2022, 22(7): 429-443. |
| [17] | SCHUSTER S, CABRERA D, ARRESE M, et al.. Triggering and resolution of inflammation in NASH[J]. Nat. Rev. Gastroenterol. Hepatol., 2018, 15(6): 349-364. |
| [18] | PARK E J, LEE J H, YU G Y, et al.. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression[J]. Cell, 2010, 140(2): 197-208. |
| [19] | PEISELER M, SCHWABE R, HAMPE J, et al.. Immune mechanisms linking metabolic injury to inflammation and fibrosis in fatty liver disease-novel insights into cellular communication circuits[J]. J. Hepatol., 2022, 77(4): 1136-1160. |
| [20] | RAPPEZ L, STADLER M, TRIANA S, et al.. SpaceM reveals metabolic states of single cells[J]. Nat. Methods, 2021, 18(7): 799-805. |
| [21] | GUILLIAMS M, BONNARDEL J, HAEST B, et al.. Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches[J]. Cell, 2022, 185(2): 379-396. |
| [22] | KISSELEVA T, BRENNER D. Molecular and cellular mechanisms of liver fibrosis and its regression[J]. Nat. Rev. Gastroenterol. Hepatol., 2021, 18(3): 151-166. |
| [23] | TSUCHIDA T, FRIEDMAN S L. Mechanisms of hepatic stellate cell activation[J]. Nat. Rev. Gastroenterol. Hepatol., 2017, 14(7): 397-411. |
| [24] | FRIEDMAN S L. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver[J]. Physiol. Rev., 2008, 88(1): 125-172. |
| [25] | TRIVEDI P, WANG S, FRIEDMAN S L. The power of plasticity-metabolic regulation of hepatic stellate cells[J]. Cell Metab., 2021, 33(2): 242-257. |
| [26] | HAMMERICH L, TACKE F. Hepatic inflammatory responses in liver fibrosis[J]. Nat. Rev. Gastroenterol. Hepatol., 2023, 20(10): 633-646. |
| [27] | CHO C S, XI J, SI Y, et al.. Microscopic examination of spatial transcriptome using Seq-Scope[J]. Cell, 2021, 184(13): 3559-3572. |
| [28] | CHUNG B K, ØGAARD J, REIMS H M, et al.. Spatial transcriptomics identifies enriched gene expression and cell types in human liver fibrosis[J]. Hepatol. Commun., 2022, 6(9): 2538-2550. |
| [29] | MATCHETT K P, WILSON-KANAMORI J R, PORTMAN J R, et al.. Multimodal decoding of human liver regeneration[J]. Nature, 2024, 630(8015): 158-165. |
| [30] | YANG J D, HAINAUT P, GORES G J, et al.. A global view of hepatocellular carcinoma: trends, risk, prevention and management[J]. Nat. Rev. Gastroenterol. Hepatol., 2019, 16(10): 589-604. |
| [31] | WU R, GUO W, QIU X, et al.. Comprehensive analysis of spatial architecture in primary liver cancer[J/OL]. Sci. Adv., 2021, 7(51): eabg3750[2025-06-24]. . |
| [32] | WU L, YAN J, BAI Y, et al.. An invasive zone in human liver cancer identified by Stereo-seq promotes hepatocyte-tumor cell crosstalk, local immunosuppression and tumor progression[J]. Cell Res., 2023, 33(8): 585-603. |
| [33] | XUE R, ZHANG Q, CAO Q, et al.. Liver tumour immune microenvironment subtypes and neutrophil heterogeneity[J]. Nature, 2022, 612(7938): 141-147. |
| [34] | YANG M, SONG X, ZHANG F, et al.. Spatial proteomic landscape of primary and relapsed hepatocellular carcinoma reveals immune escape characteristics in early relapse[J]. Hepatology, 2025, 81(5): 1452-1467. |
| [35] | LI M, WANG L, CONG L, et al.. Spatial proteomics of immune microenvironment in nonalcoholic steatohepatitis-associated hepatocellular carcinoma[J]. Hepatology, 2024, 79(3): 560-574. |
| [36] | LIU Y, XUN Z, MA K, et al.. Identification of a tumour immune barrier in the HCC microenvironment that determines the efficacy of immunotherapy[J]. J. Hepatol., 2023, 78(4): 770-782. |
| [37] | YANG Y, NI Q, LI H, et al.. Genomic and the tumor microenvironment heterogeneity in multifocal hepatocellular carcinoma[J]. Hepatology, 2024. . |
| [38] | GUO D Z, ZHANG X, ZHANG S Q, et al.. Single-cell tumor heterogeneity landscape of hepatocellular carcinoma: unraveling the pro-metastatic subtype and its interaction loop with fibroblasts[J/OL]. Mol. Cancer, 2024, 23(1): 157[2025-06-24]. . |
| [39] | CHENG Y, CHEN X, FENG L, et al.. Stromal architecture and fibroblast subpopulations with opposing effects on outcomes in hepatocellular carcinoma[J/OL]. Cell Discov., 2025, 11(1): 1[2025-06-24]. . |
| [40] | JENG W J, PAPATHEODORIDIS G V, LOK A S F. Hepatitis B[J]. Lancet, 2023, 401(10381): 1039-1052. |
| [41] | PIETSCHMANN T, BROWN R J P. Hepatitis C virus[J]. Trends Microbiol., 2019, 27(4): 379-380. |
| [42] | HSU Y C, HUANG D Q, NGUYEN M H. Global burden of hepatitis B virus: current status, missed opportunities and a call for action[J]. Nat. Rev. Gastroenterol. Hepatol., 2023, 20(8): 524-537. |
| [43] | DE SIMONE G, ANDREATA F, BLERIOT C, et al.. Identification of a Kupffer cell subset capable of reverting the T cell dysfunction induced by hepatocellular priming[J]. Immunity, 2021, 54(9): 2089-2100. |
| [44] | HENSEL N, GU Z, SAGAR, et al.. Memory-like HCV-specific CD8+ T cells retain a molecular scar after cure of chronic HCV infection[J]. Nat. Immunol., 2021, 22(2): 229-239. |
| [45] | SHI J, LI Q, LI J, et al.. Single-cell RNA sequencing reveals the spatial heterogeneity and functional alteration of endothelial cells in chronic hepatitis B infection[J/OL]. Int. J. Mol. Sci., 2024, 25(13): 7016[2025-06-24]. . |
| [46] | YU X, GONG Q, YU D, et al.. Spatial transcriptomics reveals a low extent of transcriptionally active hepatitis B virus integration in patients with HBsAg loss[J]. Gut, 2024, 73(5): 797-809. |
| [47] | CROSS A, HARRIS J M, ARBE-BARNES E, et al.. Characterisation of HBV and co-infection with HDV and HIV through spatial transcriptomics[J/OL]. Gastroenterology, 2024, 2(3): e100067[2025-06-24]. . |
| [1] | Lingfei WAN, Wenting PAN, Yuting YONG, Yuanshuai LI, Yue ZHAO, Xinlong YAN. Research Progress of Single-cell Transcriptome Sequencing Technology in Liver Fibrosis [J]. Current Biotechnology, 2024, 14(5): 793-804. |
| [2] | Shi HUANG, Yinyin MO, Lyujing LUO, Huiting LIU, Zhengyu CHEN, Genliang LI. Bioinformatics Analysis on the Regulation Mechanism of NHP2 on Hepatocellular Carcinoma Senescence [J]. Current Biotechnology, 2024, 14(1): 141-148. |
| [3] | Yanrong WANG, Yuanbiao GUO. Construction of a Prognostic Signature for Hepatocellular Carcinoma Based on Ferroptosis-related LncRNAs [J]. Current Biotechnology, 2023, 13(3): 473-481. |
| [4] | Guanqun LI, Chaojie LANG, Feng XIA. Study on Establishment and Validation of Prognostic Evaluation Model for Hepatocellular Carcinoma Based on Methylomics Database [J]. Current Biotechnology, 2023, 13(2): 282-290. |
| [5] | WANG Jiao-jiao1, GENG Yuan-yuan1, ZHANG Jia-yao2*, JIANG Wei1. Progress for Biomarkers and Detection Methods of Circulating Tumor Cell of Hepatocellular Carcinoma [J]. Curr. Biotech., 2015, 5(2): 113-119. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||