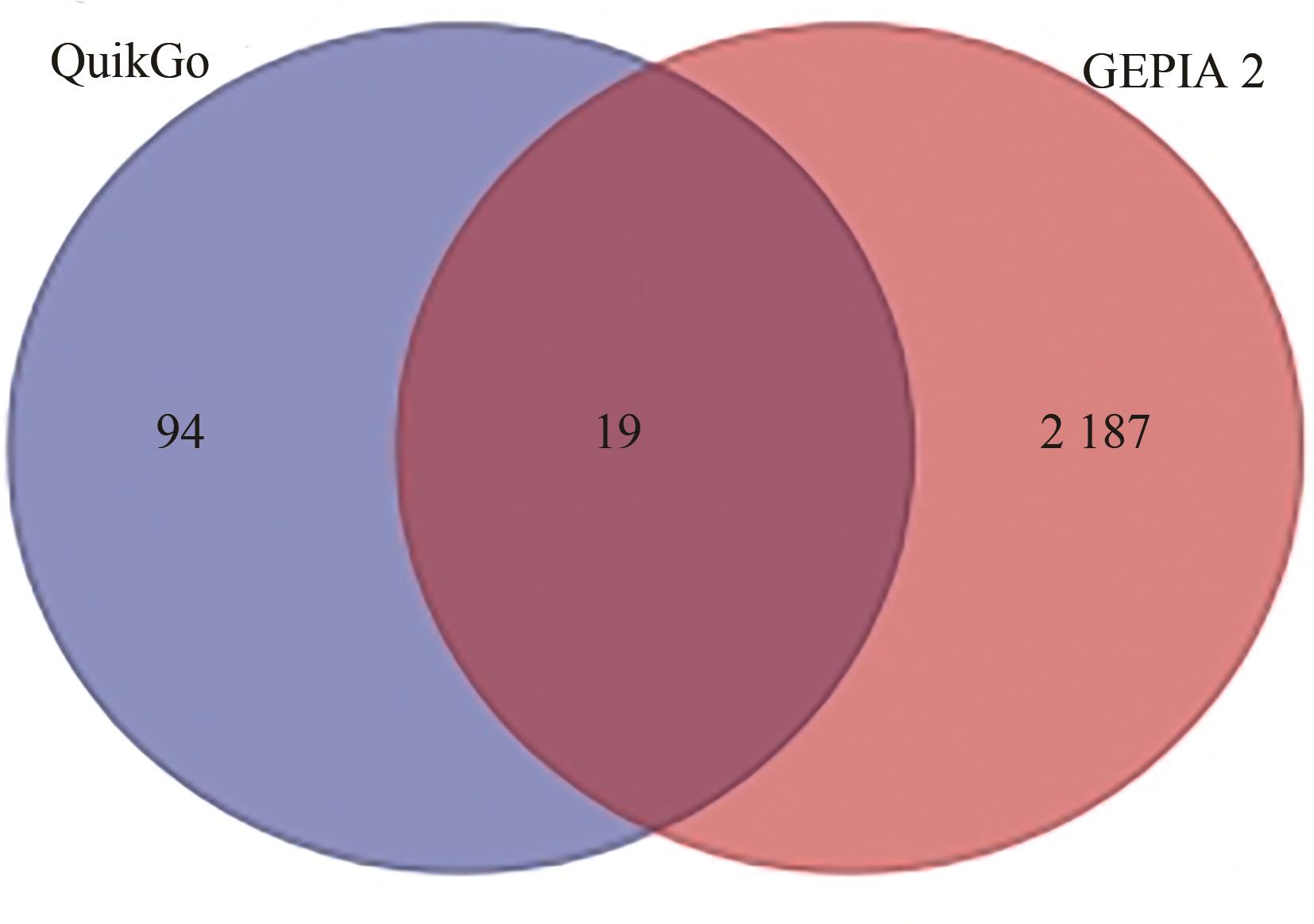
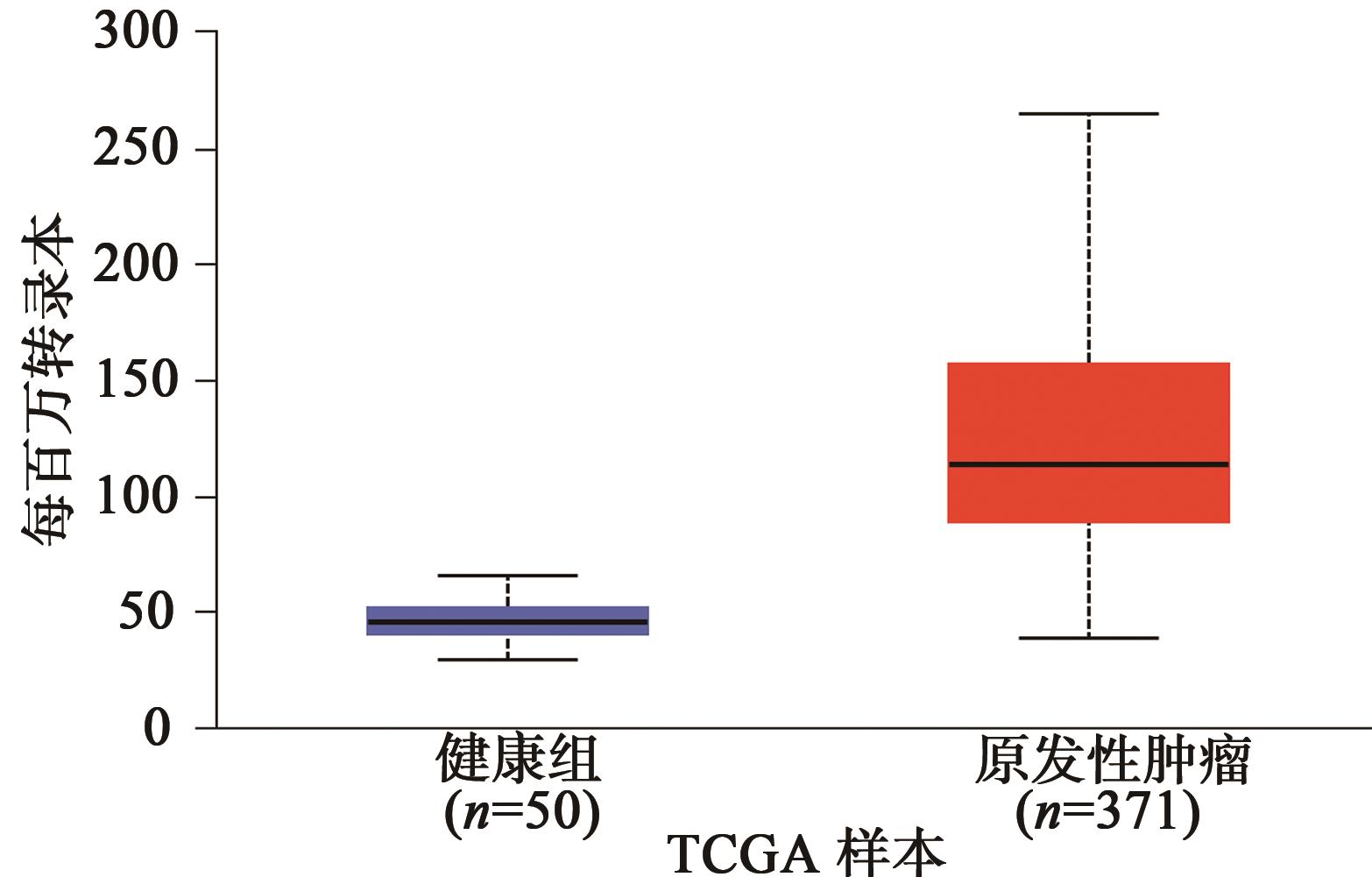
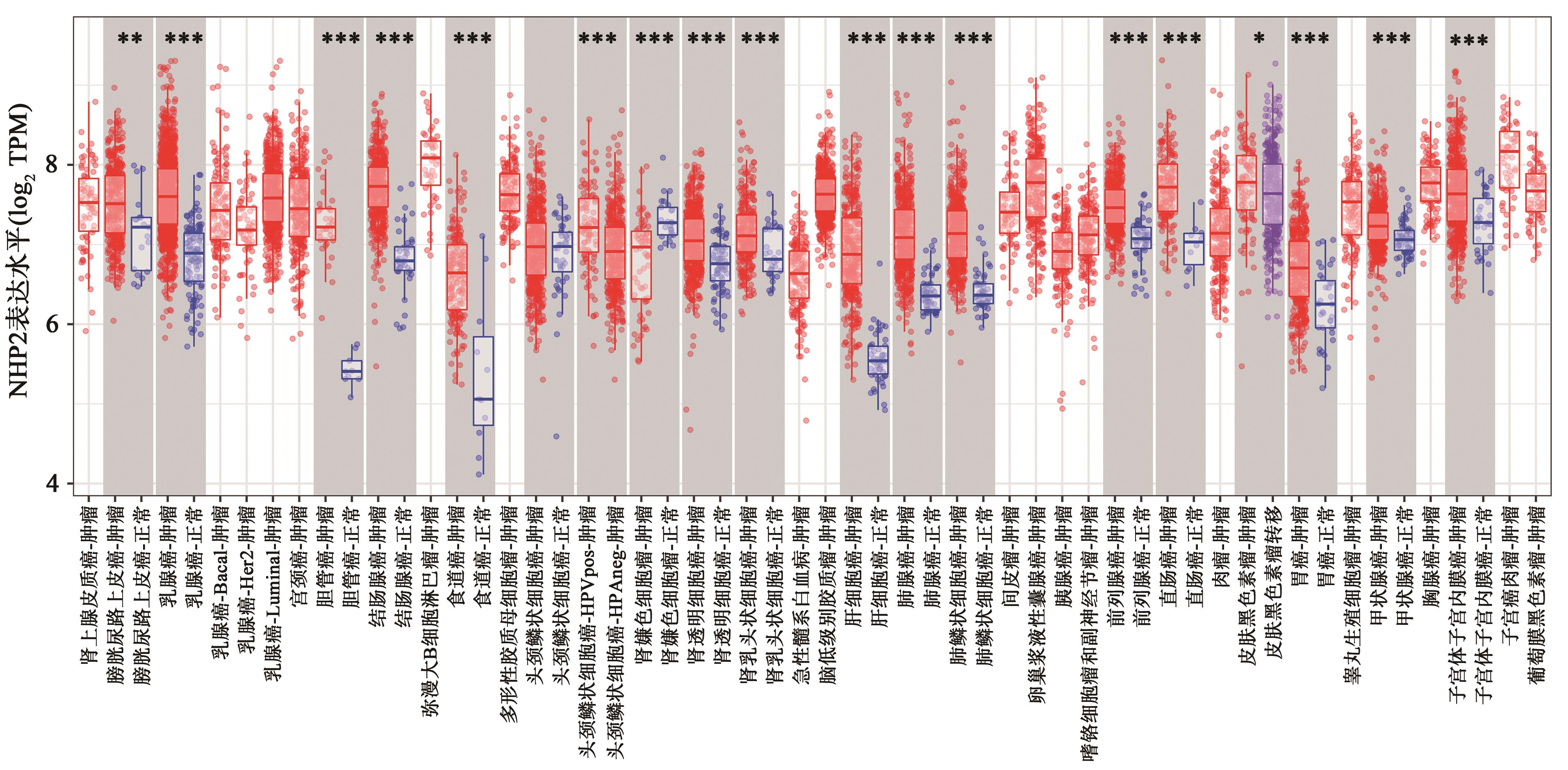
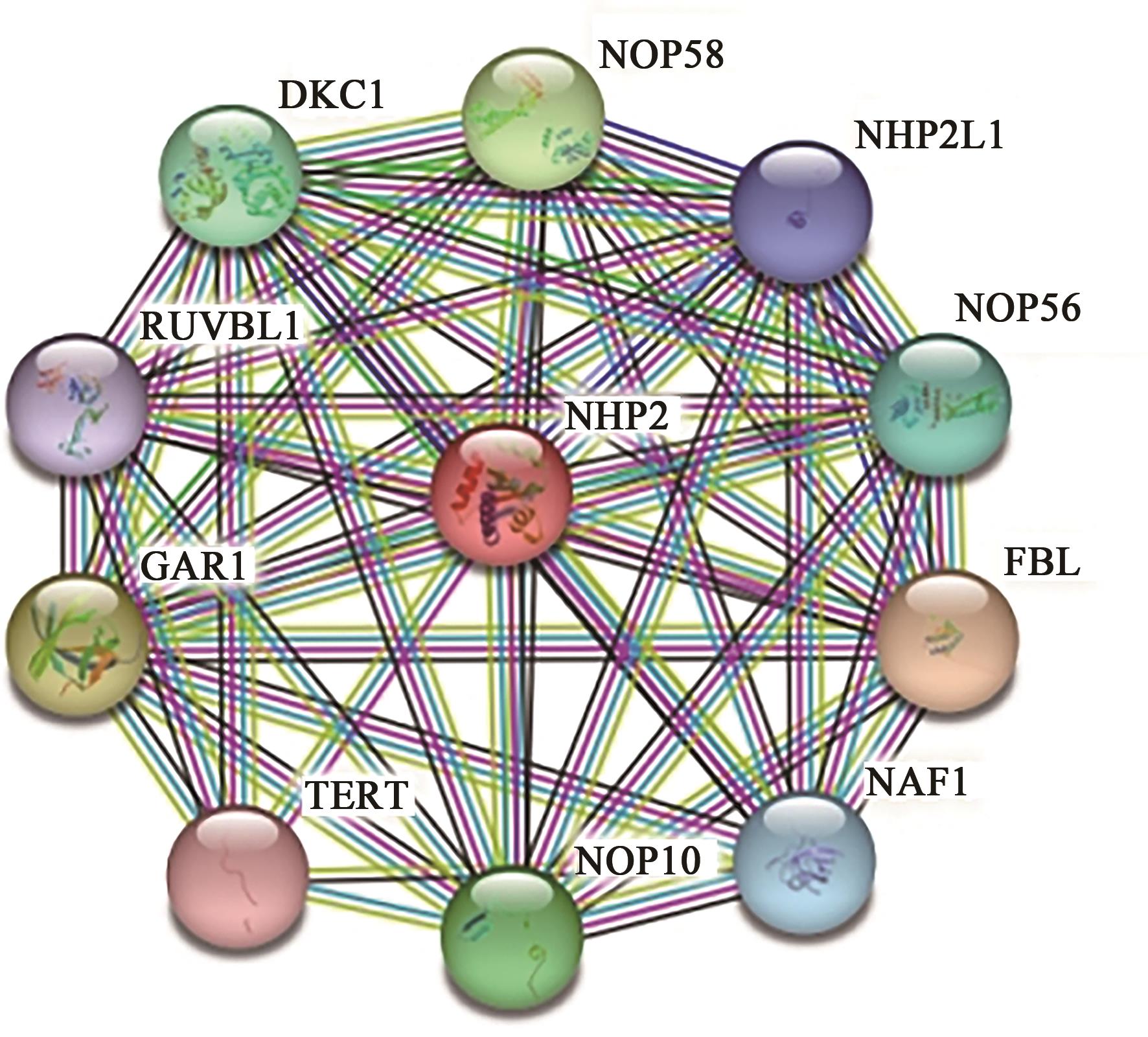
Current Biotechnology ›› 2024, Vol. 14 ›› Issue (1): 141-148.DOI: 10.19586/j.2095-2341.2023.0108
• Articles • Previous Articles Next Articles
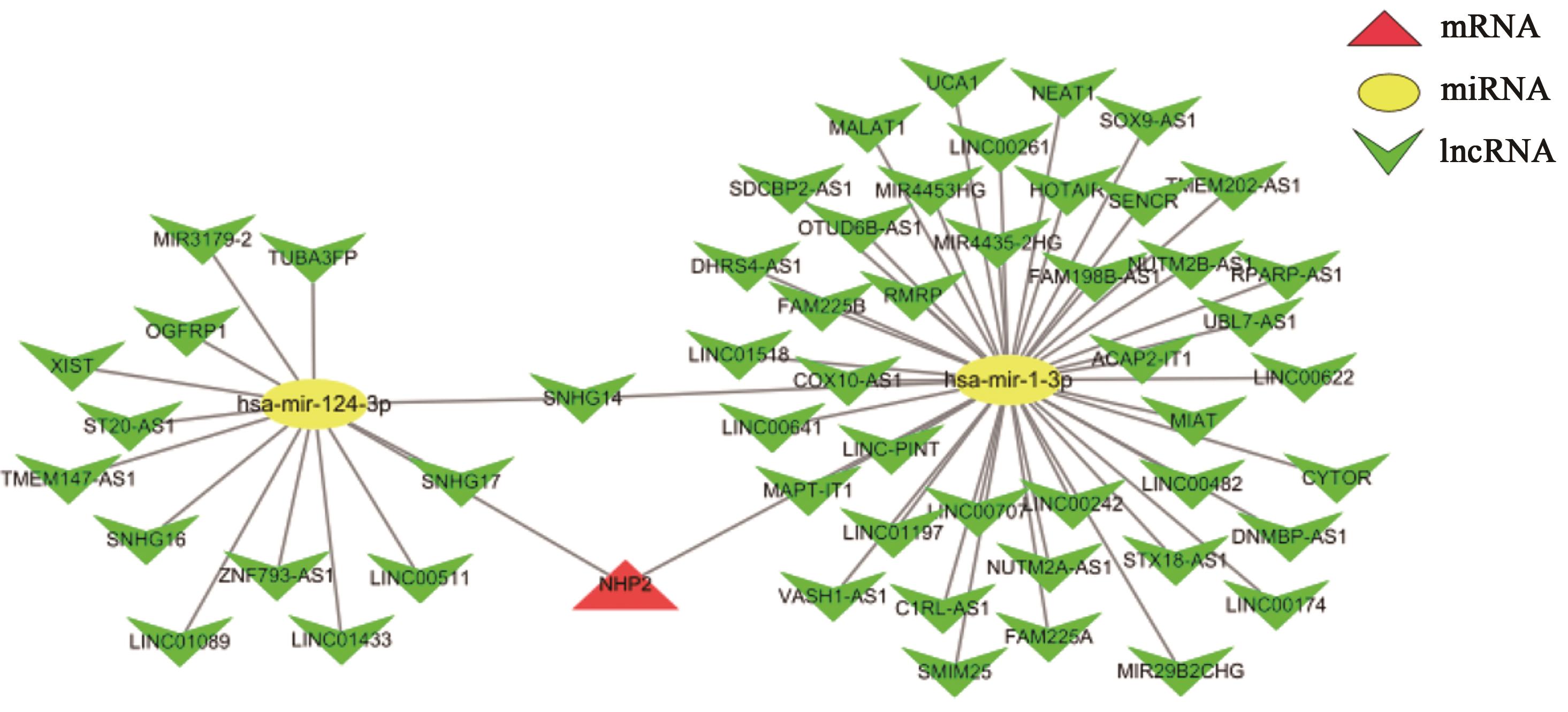
Bioinformatics Analysis on the Regulation Mechanism of NHP2 on Hepatocellular Carcinoma Senescence
Shi HUANG1,2( ), Yinyin MO1, Lyujing LUO1, Huiting LIU1, Zhengyu CHEN1, Genliang LI1(
), Yinyin MO1, Lyujing LUO1, Huiting LIU1, Zhengyu CHEN1, Genliang LI1( )
)
- 1.Youjiang Medical College for Nationalities,Guangxi Baise 533000,China
2.Affiliated Hospital of Youjiang Medical University for Nationalities,Guangxi Baise 533000,China
-
Received:2023-09-12Accepted:2023-12-08Online:2024-01-25Published:2024-02-05 -
Contact:Genliang LI
NHP2调控肝癌细胞衰老机制的生物信息学分析
黄师1,2( ), 莫茵茵1, 罗绿景1, 刘会婷1, 陈峥宇1, 李根亮1(
), 莫茵茵1, 罗绿景1, 刘会婷1, 陈峥宇1, 李根亮1( )
)
- 1.右江民族医学院,广西 百色 533000
2.右江民族医学院附属医院麻醉科,广西 百色 533000
-
通讯作者:李根亮 -
作者简介:黄师 E-mail: shihuang_mail@foxmail.com; -
基金资助:广西2023年研究生教育创新计划项目(YCSW2023502);2022年百色市科技计划项目区域多发病联合专项(百科20224148);2023年右江民族医学院硕士研究生创新项目(YXCXJH2023018)
CLC Number:
Cite this article
Shi HUANG, Yinyin MO, Lyujing LUO, Huiting LIU, Zhengyu CHEN, Genliang LI. Bioinformatics Analysis on the Regulation Mechanism of NHP2 on Hepatocellular Carcinoma Senescence[J]. Current Biotechnology, 2024, 14(1): 141-148.
黄师, 莫茵茵, 罗绿景, 刘会婷, 陈峥宇, 李根亮. NHP2调控肝癌细胞衰老机制的生物信息学分析[J]. 生物技术进展, 2024, 14(1): 141-148.
share this article
| 1 | 秦叔逵,任正刚,曾昭冲,等.原发性肝癌诊疗指南(2022年版)[J].肿瘤综合治疗电子杂志,2022,8(2):16-53. |
| QIN S, REN Z, ZENG Z,et al..Guidelines for diagnosis and treatment of primary liver cancer (2022 edition)[J]. J. Multidiscip. Cancer Manag. Electron. Version, 2022, 8(2): 16-53. | |
| 2 | 雷倩,夏机良,冯湘玲,等.NEK2通过抵抗肝癌细胞衰老促进肝癌进展[J].中南大学学报(医学版),2022,47(2):153-164. |
| LEI Q, XIA J L, FENG X L, et al.. NEK2 promotes the progression of liver cancer by resisting the cellular senescence[J]. J. Cent. South Univ. Med. Sci., 2022, 47(2): 153-164. | |
| 3 | ROAKE C M, ARTANDI S E. Regulation of human telomerase in homeostasis and disease[J]. Nat. Rev. Mol. Cell Biol., 2020, 21(7): 384-397. |
| 4 | RAGHUNANDAN M, GEELEN D, MAJEROVA E, et al.. NHP2 downregulation counteracts hTR-mediated activation of the DNA damage response at ALT telomeres[J/OL]. EMBO J., 2021, 40(6): e106336[2023-11-20]. . |
| 5 | MALIŃSKI B, VERTEMARA J, FAUSTINI E, et al.. Novel pathological variants of NHP2 affect N-terminal domain flexibility, protein stability, H/ACA Ribonucleoprotein (RNP) complex formation and telomerase activity[J]. Hum. Mol. Genet., 2023, 32(19): 2901-2912. |
| 6 | LIU L, SHENG Y, WANG C Y, et al.. A novel mutation (p.Y24N) inNHP2 leads to idiopathic pulmonary fibrosis and lung carcinoma chronic obstructive lung disease by disrupting the expression and nucleocytoplasmic localization of NHP2[J/OL]. Biochim. Biophys. Acta Mol. Basis. Dis., 2023, 1869(5): 166692[2023-11-20]. . |
| 7 | SUNG H, FERLAY J, SIEGEL R L, et al.. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J. Clin., 2021, 71(3): 209-249. |
| 8 | 黄家利,徐畅,吴虹,等.肝细胞癌中HP1BP3的表达及其对细胞恶性生物学行为的影响[J].临床与实验病理学杂志,2022,38(6):653-658. |
| HUANG J L, XU C, WU H, et al.. Expression of HP1 BP3 in hepatocellular carcinoma and its effect on malignant biological behaviors of the tumor cells[J]. Chin. J. Clin. Exp. Pathol., 2022, 38(6): 653-658. | |
| 9 | 陶莹,廖长秀.新兴免疫阻断分子在肝细胞癌发生发展中的作用[J].临床肝胆病杂志,2023,39(4):948-955. |
| TAO Y, LIAO C X. Role of new potential immune blocking molecules in the development and progression of hepatocellular carcinoma[J]. J. Clin. Hepatol., 2023, 39(4): 948-955. | |
| 10 | 崔晓东,邹娇娇,王转花.细胞衰老与癌症治疗[J].中国生物化学与分子生物学报,2020,36(2):119-126. |
| CUI X D, ZOU J J, WANG Z H. Cell senescence and cancer therapy[J]. Chin. J. Biochem. Mol. Biol., 2020, 36(2): 119-126. | |
| 11 | CAO H, ZHAI Y, JI X, et al.. Noncoding telomeric repeat-containing RNA inhibits the progression of hepatocellular carcinoma by regulating telomerase-mediated telomere length[J]. Cancer Sci., 2020, 111(8): 2789-2802. |
| 12 | TANG S, WU W, WAN H, et al.. Knockdown of NHP2 inhibits hepatitis B virus X protein-induced hepatocarcinogenesis via repressing TERT expression and disrupting the stability of telomerase complex[J]. Aging Albany NY, 2020, 12(19): 19365-19374. |
| 13 | NAULT J C. Pathogenesis of hepatocellular carcinoma according to aetiology[J]. Best Pract. Res. Clin. Gastroenterol., 2014, 28(5): 937-947. |
| 14 | KIM Y J, YOO J E, JEON Y, et al.. Suppression of PROX1-mediated TERT expression in hepatitis B viral hepatocellular carcinoma[J]. Int. J. Cancer, 2018, 143(12): 3155-3168. |
| 15 | JAISWAL R K, YADAVA P K. Assessment of telomerase as drug target in breast cancer[J/OL]. J. Biosci., 2020, 45(1): 72[2023-11-20]. . |
| 16 | DOGAN F, BIRAY AVCI C. Correlation between telomerase and mTOR pathway in cancer stem cells[J]. Gene, 2018, 641: 235-239. |
| 17 | DRATWA M, WYSOCZAŃSKA B, ŁACINA P, et al.. TERT-regulation and roles in cancer formation[J/OL]. Immunol, 2020, 11: 589929[2023-11-20]. . |
| 18 | LI Y, TERGAONKAR V. Telomerase reactivation in cancers: mechanisms that govern transcriptional activation of the wild-type vs. mutant TERT promoters[J]. Transcription, 2016, 7(2): 44-49. |
| 19 | ZHEN H, DU P, YI Q, et al.. LINC00958 promotes bladder cancer carcinogenesis by targeting miR-490-3p and AURKA[J/OL]. BMC Cancer, 2021, 21(1): 1145[2023-11-20]. . |
| 20 | HUANG C S, TSAI C H, YU C P, et al.. Long noncoding RNA LINC02470 sponges microRNA-143-3p and enhances SMAD3-mediated epithelial-to-mesenchymal transition to promote the aggressive properties of bladder cancer[J/OL]. Cancers Basel, 2022, 14(4): 968[2023-11-20]. . |
| 21 | 吴潇芸,吴林秀,张丽娣,等.LINC00839靶向调控miR-124-3p对膀胱癌细胞生物学行为的影响[J].天津医药,2023,51(5):464-468. |
| WU X Y, WU L X, ZHANG L D, et al.. The effect of LINC00839 targeting regulation of miR-124-3p on biological behaviors of bladder cancer cells[J]. Tianjin Med. J., 2023, 51(5): 464-468. | |
| 22 | 苏敏,王文祥,唐金明.circHIPK3通过调控miR-124-3p/EZH2促进食管鳞状细胞癌细胞恶性生物学行为[J].临床与实验病理学杂志,2021,37(12):1405-1411. |
| SU M, WANG W X, TANG J M. CircHIPK3 promotes malignant biological behavior of esophageal squamous cell carcinoma cells through regulation of miR-124-3p/EZH2[J]. Chin. J. Clin. Exp. Pathol., 2021, 37(12): 1405-1411. | |
| 23 | 周立飞,高跃丽,耿欣,等. MiR-124-3p和SEPT9与宫颈癌患者临床表现的相关性分析[J].中国优生与遗传杂志,2023,31(1):64-69. |
| ZHOU L F, GAO Y L, GENG X, et al.. Correlation analysis of miR-124-3p and SEPT9 with clinical manifestations of cervical cancer patients[J]. Chin. J. Birth Health Hered., 2023, 31(1): 64-69. | |
| 24 | 李敬霞,刘艳.LncRNA PTPRG-AS1通过靶向miR-124-3p调控肝癌细胞凋亡和放射敏感性的研究机制[J].现代肿瘤医学,2021,29(10):1676-1682. |
| LI J X, LIU Y. LncRNA PTPRG-AS1 regulates apoptosis and radiosensitivity of hepatoma cells by targeting miR-124-3p [J]. J. Mod. Oncol., 2021, 29(10): 1676-1682. | |
| 25 | WU Q, ZHONG H, JIAO L, et al.. MiR-124-3p inhibits the migration and invasion of Gastric cancer by targeting ITGB3[J/OL]. Pathol. Res. Pract., 2020, 216(1): 152762[2023-11-20]. . |
| 26 | ZHAO J T, CHI B J, SUN Y, et al.. LINC00174 is an oncogenic lncRNA of hepatocellular carcinoma and regulates miR-320/S100A10 axis[J]. Cell Biochem. Funct., 2020, 38(7): 859-869. |
| 27 | 张英英.lncRNA CYTOR、miRNA378a与FEN1基因共同调控肝癌细胞恶性生物学行为的机制研究[D].郑州:郑州大学,2020. |
| [1] | Lingfei WAN, Wenting PAN, Yuting YONG, Yuanshuai LI, yue ZHAO, Xinlong YAN. Research Progress in Spatial Transcriptomics Technology for Liver Disease Research [J]. Current Biotechnology, 2025, 15(4): 645-654. |
| [2] | Man LIU, Yongxi HUANG, Pupu YAN, Jiali LIU, Liwei GUO. Identification of Shared Genetic Characteristics and Biological Mechanisms of Newcastle Disease and Streptococcal Disease in Chickens Based on the GEO Database [J]. Current Biotechnology, 2024, 14(3): 451-458. |
| [3] | Wenran HU, Xiaoyan HAO, Zhun ZHAO, Wukui SHAO, Shengqi GAO, Jianping LI, Quansheng HUANG. Bioinformatic Analysis of the CAD Gene Family in Cotton [J]. Current Biotechnology, 2023, 13(3): 412-424. |
| [4] | Yanrong WANG, Yuanbiao GUO. Construction of a Prognostic Signature for Hepatocellular Carcinoma Based on Ferroptosis-related LncRNAs [J]. Current Biotechnology, 2023, 13(3): 473-481. |
| [5] | Guanqun LI, Chaojie LANG, Feng XIA. Study on Establishment and Validation of Prognostic Evaluation Model for Hepatocellular Carcinoma Based on Methylomics Database [J]. Current Biotechnology, 2023, 13(2): 282-290. |
| [6] | Xingshu ZHU, Junjie HUANG, Caiming WENG, Wangwu LIU, Yongyuan CHEN, Huiyan CHEN, Hu ZHAO, Yu WANG, Chengzhi LIN. Screening and Functional Enrichment Analysis of Key Hub Genes in Lynch Syndrome-associated Colorectal Cancer Based on Bioinformatics Analysis [J]. Current Biotechnology, 2023, 13(2): 291-297. |
| [7] | Fengmei LI, Xueyuan TANG, Gaoyang JIAO, Yuting QI, Xianting DU, Qianqian BAO, Meng ZHANG, Xiaobo XU, Xin XU, Yanfang ZHANG. Identification and Analysis of Genes Controlling Flower Shape in Lonicerajaponica Thunb. [J]. Current Biotechnology, 2023, 13(1): 90-101. |
| [8] | Min YU, Min WANG, Yanhuan WEI, Yiyi LIU. Analysis of Potential Key Molecular Biomarkers and Immune Infiltration Characteristics of SARS-CoV-2 Virus Infection [J]. Current Biotechnology, 2022, 12(5): 760-768. |
| [9] | XU Guangping, ZHANG Zhiyong, REN Zhonghong, ZHANG Zhiwei*. Bioinformatics Analysis of the Structure and Function of the Protein Encoded by ELO Gene in Acanthopagrus schlegelii [J]. Curr. Biotech., 2021, 11(3): 344-352. |
| [10] | XU Jie, WANG Kefei, WEI Xiaojing, GONG Lixin, JIAO Yang, QIU Lugui, HAO Mu*. Expression and Prognostic Significance of SCHIP1 in Acute Myeloid Leukemia: Analysis Based on Bioinformatics [J]. Curr. Biotech., 2020, 10(4): 417-425. |
| [11] | ZHANG Fengli1, YANG Yalin1*, XIA Rui1, GAO Chenchen1, LI Dong1, RAN Chao1, ZHANG Zhen2, ZHANG Hongling2, ZHOU Zhigang2*. Gene Cloning and Bioinformatics Analysis of Secret Protease Gene from Thelohanellus kitauei [J]. Curr. Biotech., 2019, 9(4): 375-383. |
| [12] | XU Changlu, ZHAO Yanhong, MA Yige, WANG Bingrui, WANG Ding, GUO Qing, TONG Jingyuan, GAO Jie, LI Yapu, LIU Jinhua, SHI Lihong*. Dissecting the Dynamic Expression of Autophagy Related Genes at Each Stage of Human Terminal Erythriod Differentiation Through Bioinformatic Methods [J]. Curr. Biotech., 2019, 9(3): 271-276. |
| [13] | SUN Jingjuan1, QIU Jingxuan1, ZENG Haijuan1, DING Chengchao1, WANG Guangbin2, LI Jie1, WANG Shujuan1, LIU Qing1*. Truncated Protein Expression, Purification and Polyclonal Antibody Preparation of PrsA1 Gene from Listeria monocytogenes [J]. Curr. Biotech., 2018, 8(3): 254-260. |
| [14] | JIANG Zhilei, ZHOU Lin, WU Fengci, SONG Xinyuan*. Analysis of Location and Expression Pattern of Maize Expansin Gene Family [J]. Curr. Biotech., 2018, 8(1): 34-40. |
| [15] | ZHANG Maona, JIANG Liang*, ZHANG Yan*. Advance on Bioinformatic Analysis of Selenium Metabolic Network and Selenoproteomes [J]. Curr. Biotech., 2017, 7(5): 537-544. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||