Current Biotechnology ›› 2024, Vol. 14 ›› Issue (2): 237-247.DOI: 10.19586/j.2095-2341.2023.0133
• Articles • Previous Articles Next Articles
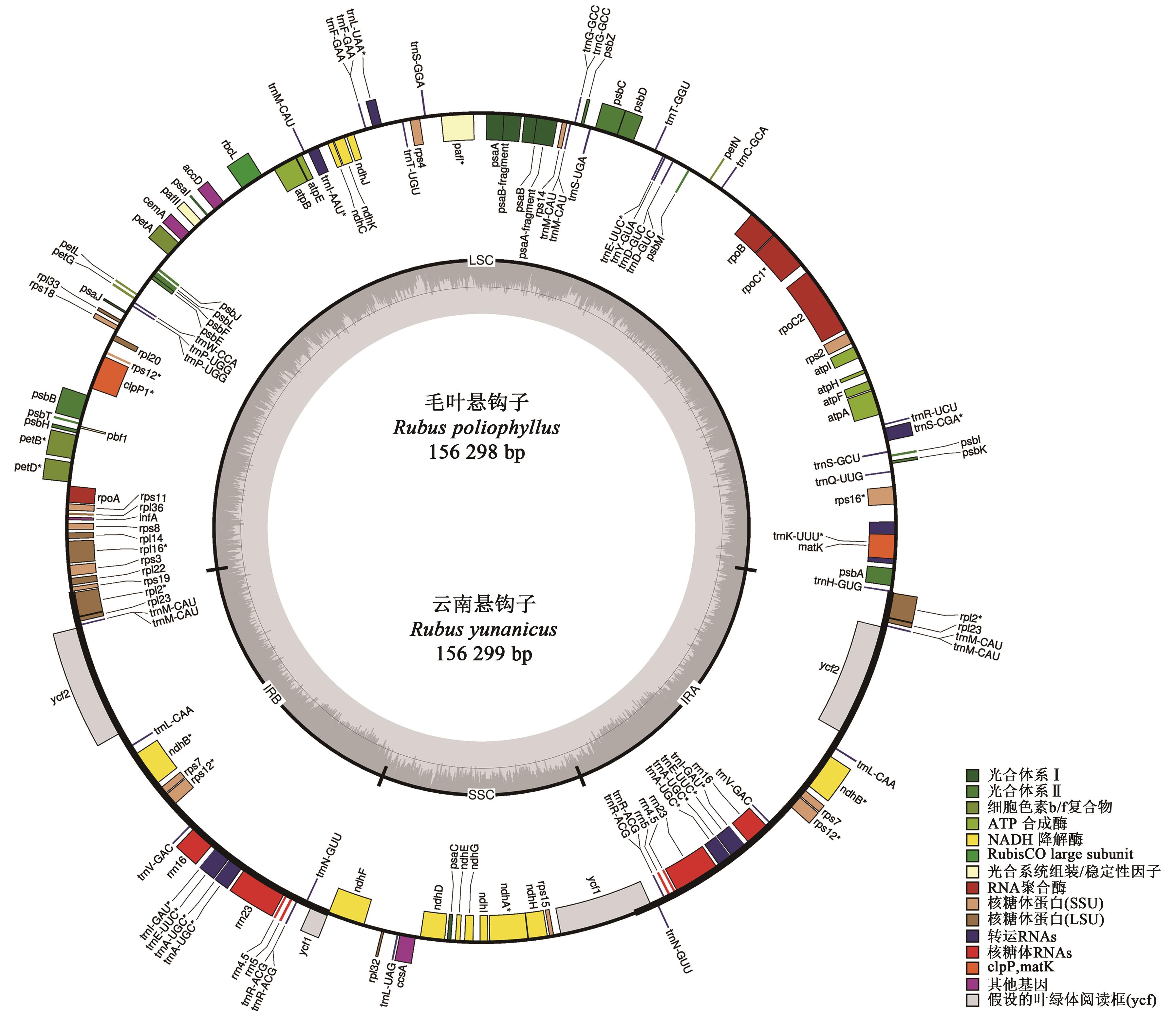
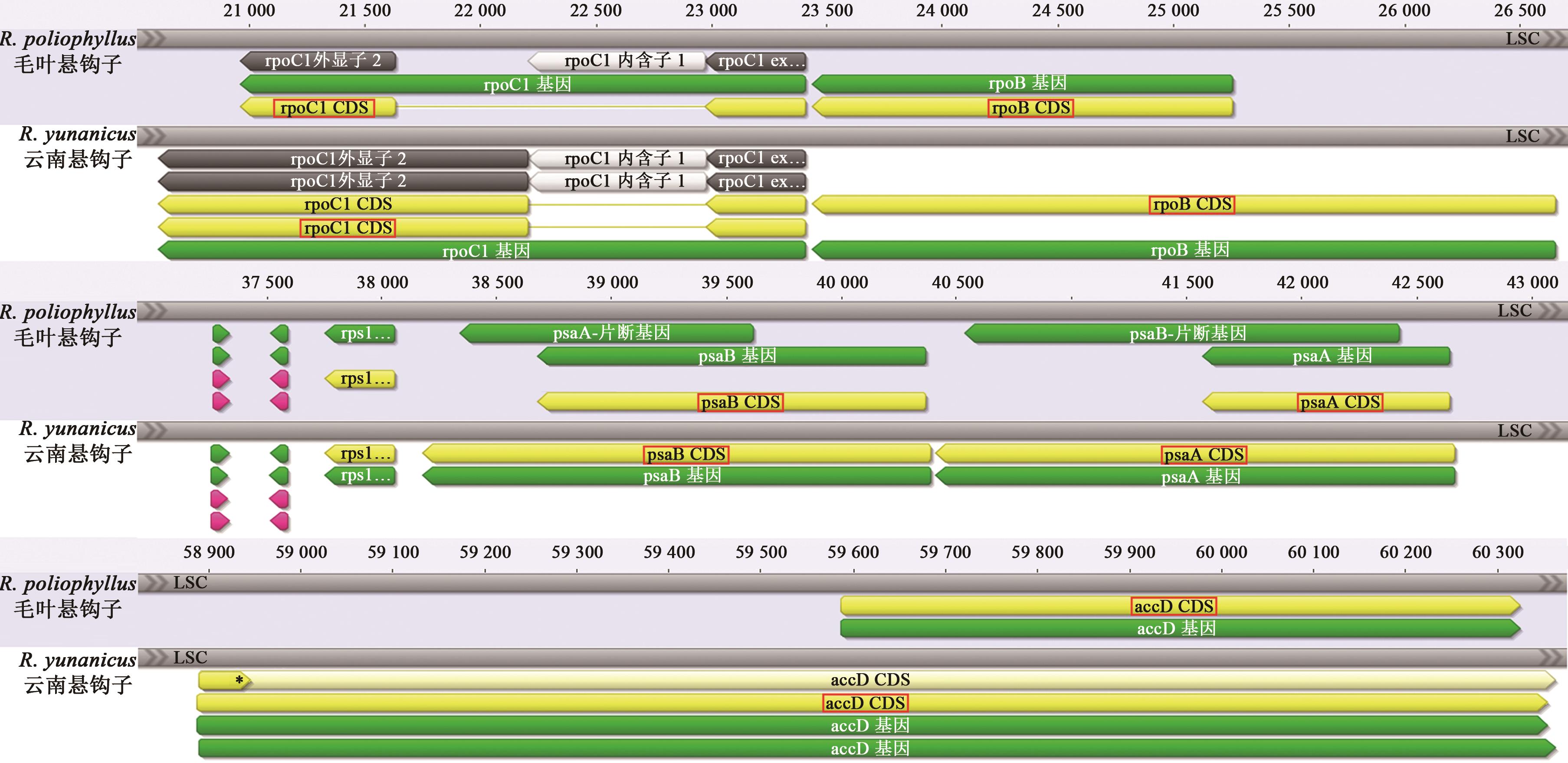
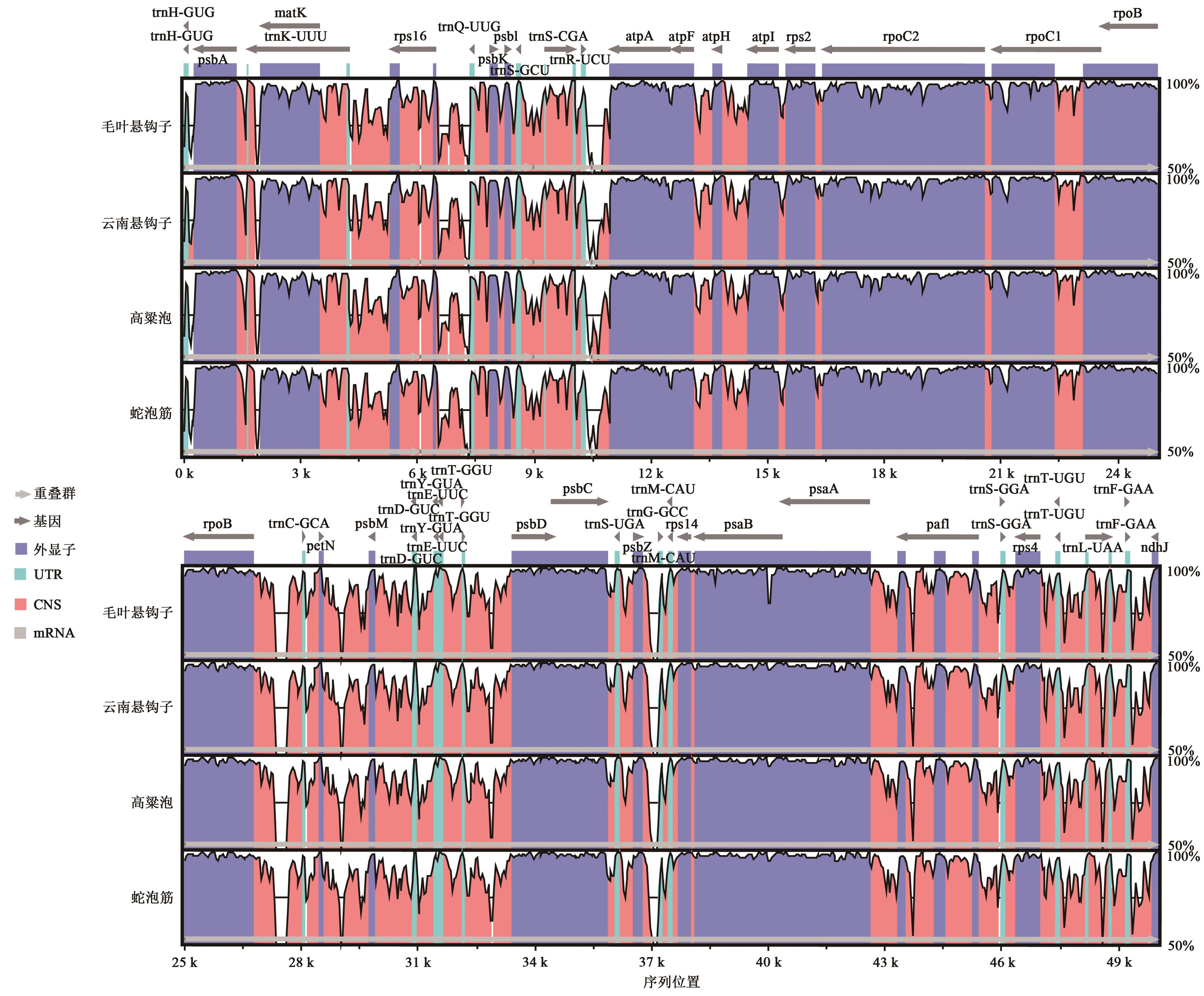
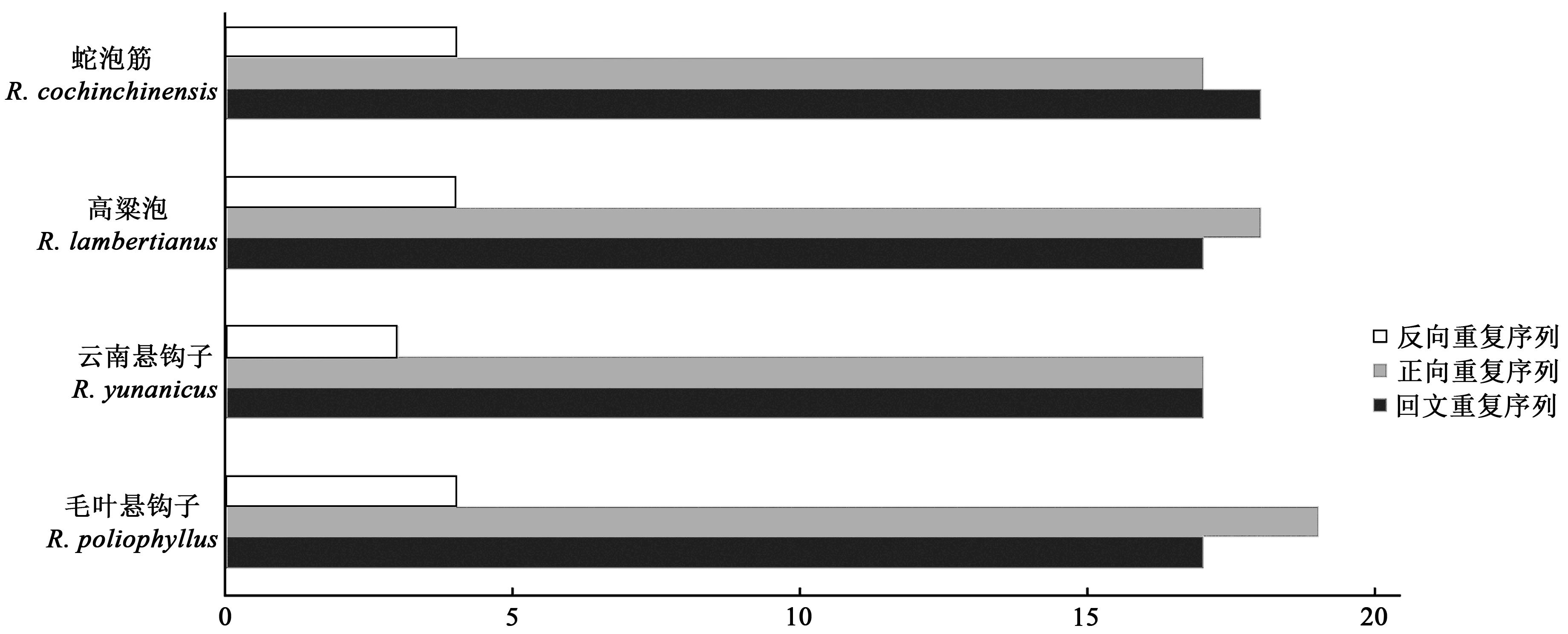
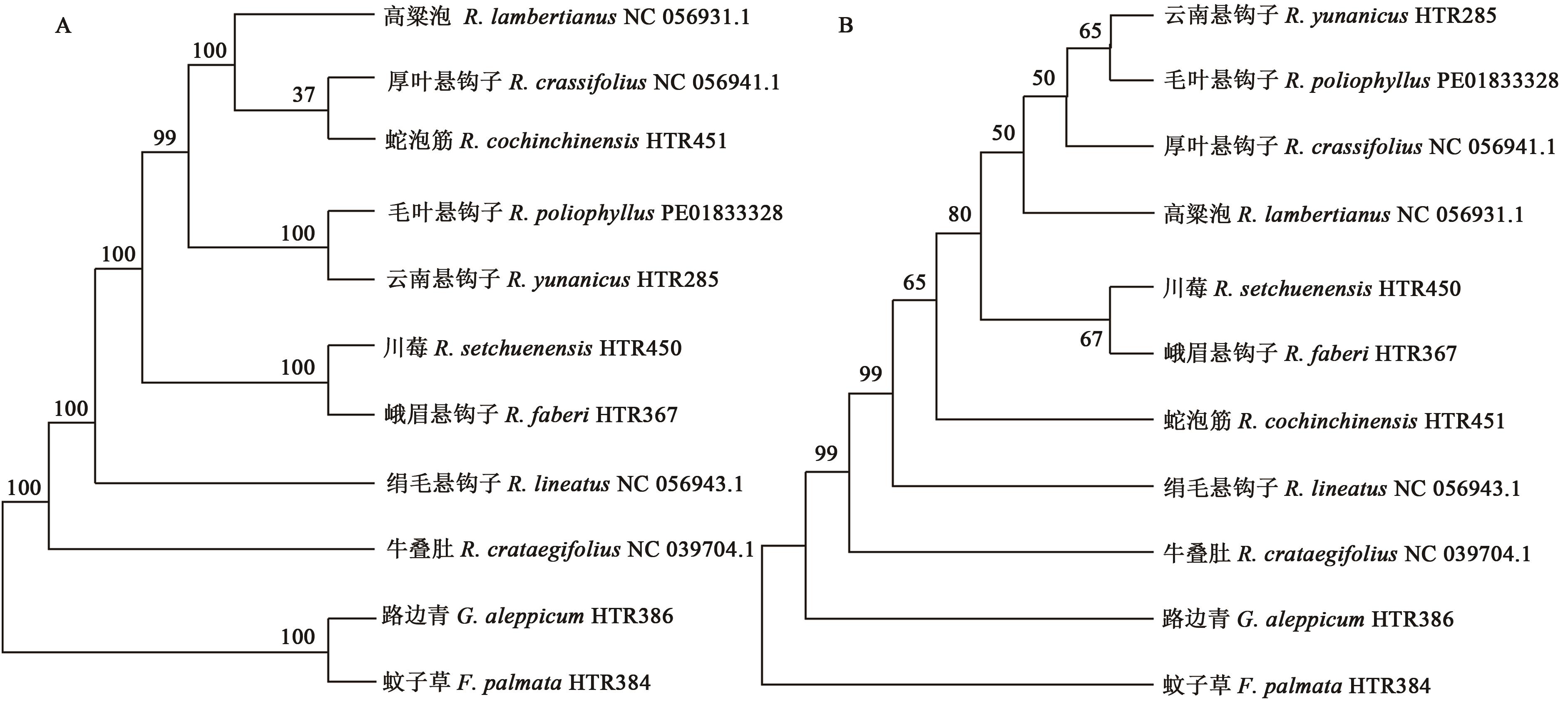
Comparative Analysis of Chloroplast Genomes from Rubuspoliophyllus and Rubus yunanicus
Kang HU1,2( ), Liping YU1,2, Wei GU1, Zhimin HE1,2, Junxin ZHANG1,2, Zixuan LIU1,2, Tiran HUANG1,2,3(
), Liping YU1,2, Wei GU1, Zhimin HE1,2, Junxin ZHANG1,2, Zixuan LIU1,2, Tiran HUANG1,2,3( ), Mingfeng YANG1,2(
), Mingfeng YANG1,2( )
)
- 1.College of Biological and Resource Environment,Beijing University of Agriculture,Beijing 102206,China
2.Key Laboratory for Northern Urban Agriculture of Ministry of Agriculture and Rural Affairs,Beijing University of Agriculture,Beijing 102206,China
3.College of Life Sciences,University of Chinese Academy of Sciences,Beijing 100049,China
-
Received:2023-10-23Accepted:2023-11-29Online:2024-03-25Published:2024-04-17 -
Contact:Tiran HUANG,Mingfeng YANG
毛叶悬钩子和云南悬钩子叶绿体基因组比较分析
胡康1,2( ), 于丽平1,2, 谷薇1, 贺志敏1,2, 张峻鑫1,2, 刘子璇1,2, 黄体冉1,2,3(
), 于丽平1,2, 谷薇1, 贺志敏1,2, 张峻鑫1,2, 刘子璇1,2, 黄体冉1,2,3( ), 杨明峰1,2(
), 杨明峰1,2( )
)
- 1.北京农学院生物与资源环境学院,北京 102206
2.北京农学院,农业农村部华北都市农业重点实验室,北京 102206
3.中国科学院大学生命科学学院,北京 100049
-
通讯作者:黄体冉,杨明峰 -
作者简介:胡康 E-mail: gan9933@163.com -
基金资助:北京市属高等学校高水平教学创新团队建设支持计划项目(BPHR20220211);北京农学院2020年和2021年本科生科研训练项目
CLC Number:
Cite this article
Kang HU, Liping YU, Wei GU, Zhimin HE, Junxin ZHANG, Zixuan LIU, Tiran HUANG, Mingfeng YANG. Comparative Analysis of Chloroplast Genomes from Rubuspoliophyllus and Rubus yunanicus[J]. Current Biotechnology, 2024, 14(2): 237-247.
胡康, 于丽平, 谷薇, 贺志敏, 张峻鑫, 刘子璇, 黄体冉, 杨明峰. 毛叶悬钩子和云南悬钩子叶绿体基因组比较分析[J]. 生物技术进展, 2024, 14(2): 237-247.
share this article
| 物种 | 长度/bp | GC含量/% | rRNA | tRNA | 内含子基因 | 编码序列 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cp | LSC | SSC | IRa | IRb | Cp | LSC | SSC | IR | |||||
| 毛叶悬钩子(R. poliophyllus) | 156 298 | 85 894 | 18 850 | 25 777 | 25 777 | 37.2 | 35.1 | 31.2 | 42.8 | 10 | 45 | 27 | 104 |
| 云南悬钩子(R. yunanicus) | 156 299 | 85 896 | 18 849 | 25 777 | 25 777 | 37.2 | 35.1 | 31.2 | 42.8 | 10 | 45 | 27 | 104 |
| 高粱泡(R. lambertianus) | 156 316 | 85 879 | 18 873 | 25 782 | 25 782 | 37.2 | 35.1 | 31.2 | 42.8 | 10 | 45 | 27 | 104 |
| 高粱泡变种(R. lambertianus var.) | 156 569 | 86 123 | 18 874 | 25 782 | 28 790 | 37.2 | 35.1 | 31.2 | 42.8 | 10 | 45 | 27 | 104 |
| 蛇泡筋(R. cochinchinensis) | 156 240 | 85 850 | 18 848 | 25 771 | 25 771 | 37.2 | 35.1 | 31.2 | 42.8 | 10 | 45 | 27 | 104 |
Table 1 Annotated statistical table of chloroplast genomes of Rubus plants
| 物种 | 长度/bp | GC含量/% | rRNA | tRNA | 内含子基因 | 编码序列 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cp | LSC | SSC | IRa | IRb | Cp | LSC | SSC | IR | |||||
| 毛叶悬钩子(R. poliophyllus) | 156 298 | 85 894 | 18 850 | 25 777 | 25 777 | 37.2 | 35.1 | 31.2 | 42.8 | 10 | 45 | 27 | 104 |
| 云南悬钩子(R. yunanicus) | 156 299 | 85 896 | 18 849 | 25 777 | 25 777 | 37.2 | 35.1 | 31.2 | 42.8 | 10 | 45 | 27 | 104 |
| 高粱泡(R. lambertianus) | 156 316 | 85 879 | 18 873 | 25 782 | 25 782 | 37.2 | 35.1 | 31.2 | 42.8 | 10 | 45 | 27 | 104 |
| 高粱泡变种(R. lambertianus var.) | 156 569 | 86 123 | 18 874 | 25 782 | 28 790 | 37.2 | 35.1 | 31.2 | 42.8 | 10 | 45 | 27 | 104 |
| 蛇泡筋(R. cochinchinensis) | 156 240 | 85 850 | 18 848 | 25 771 | 25 771 | 37.2 | 35.1 | 31.2 | 42.8 | 10 | 45 | 27 | 104 |
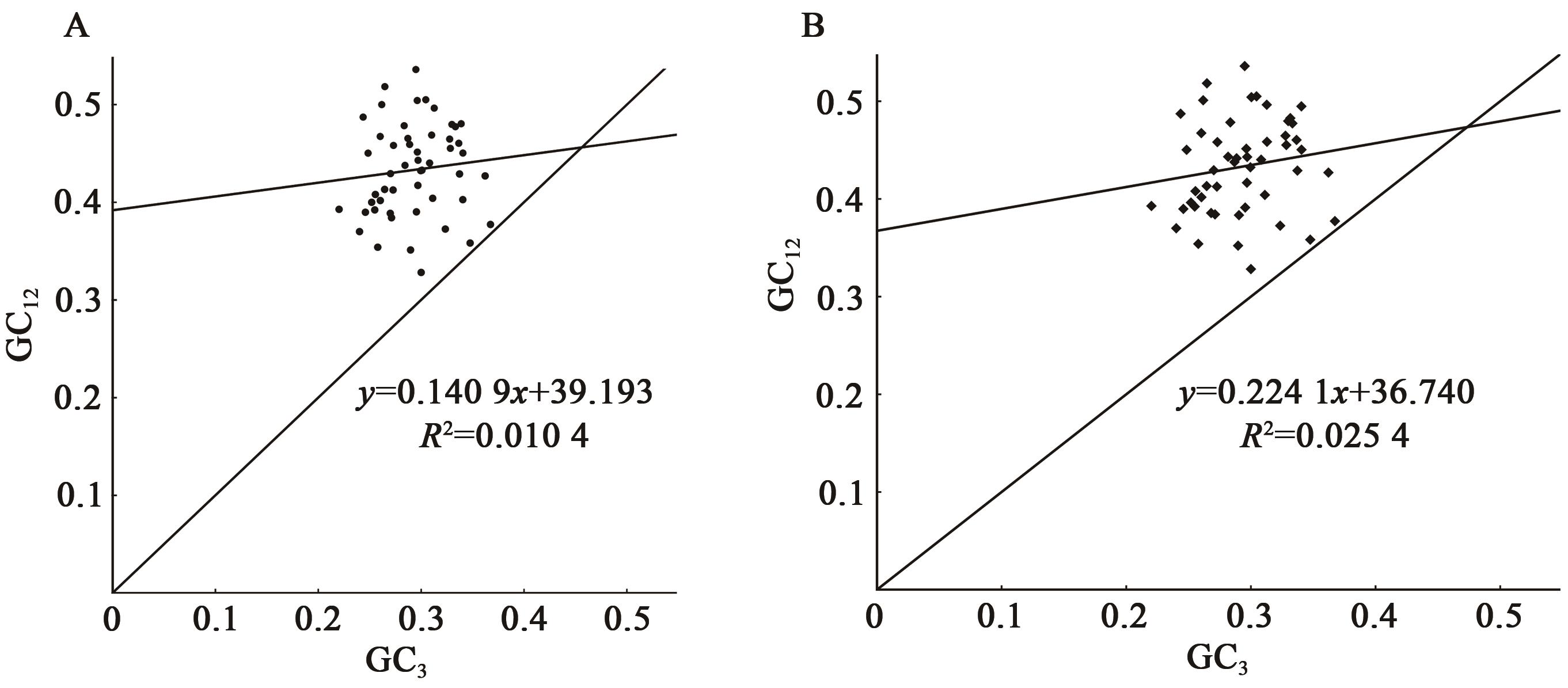
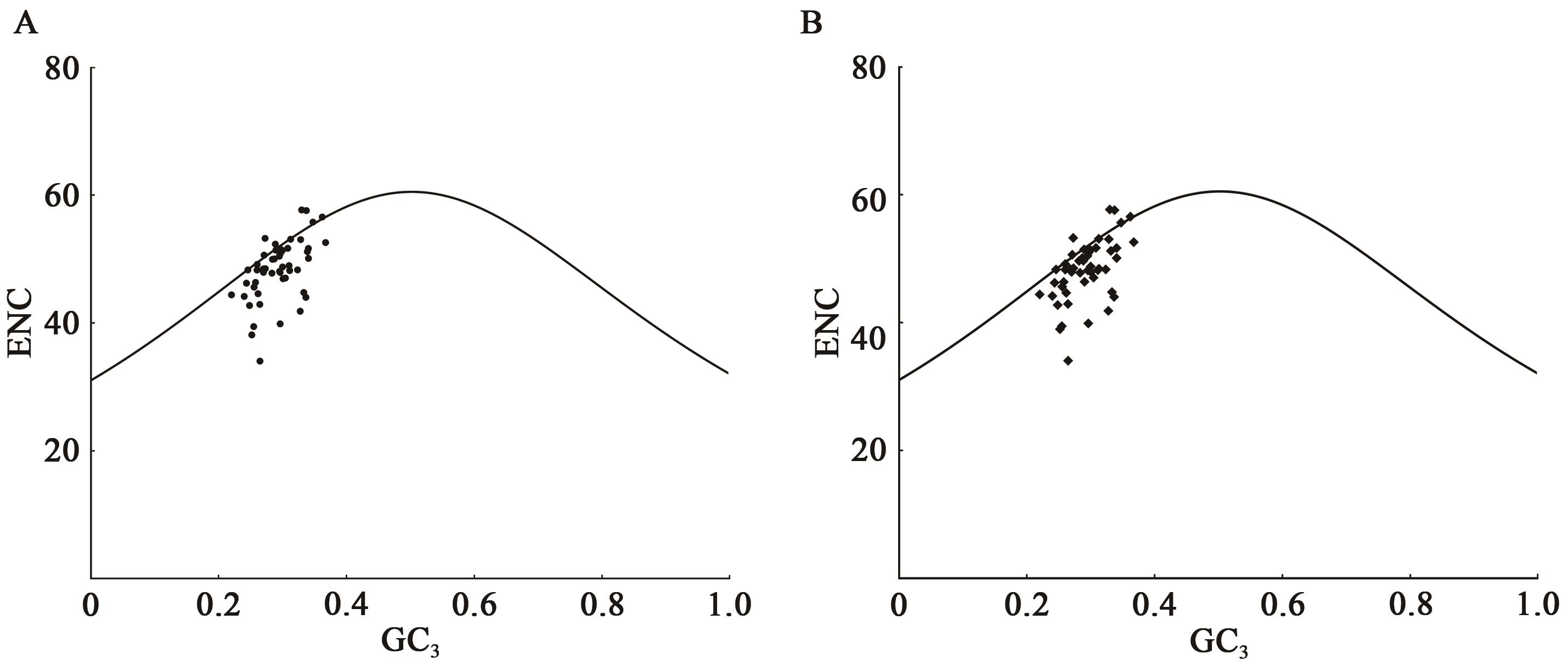
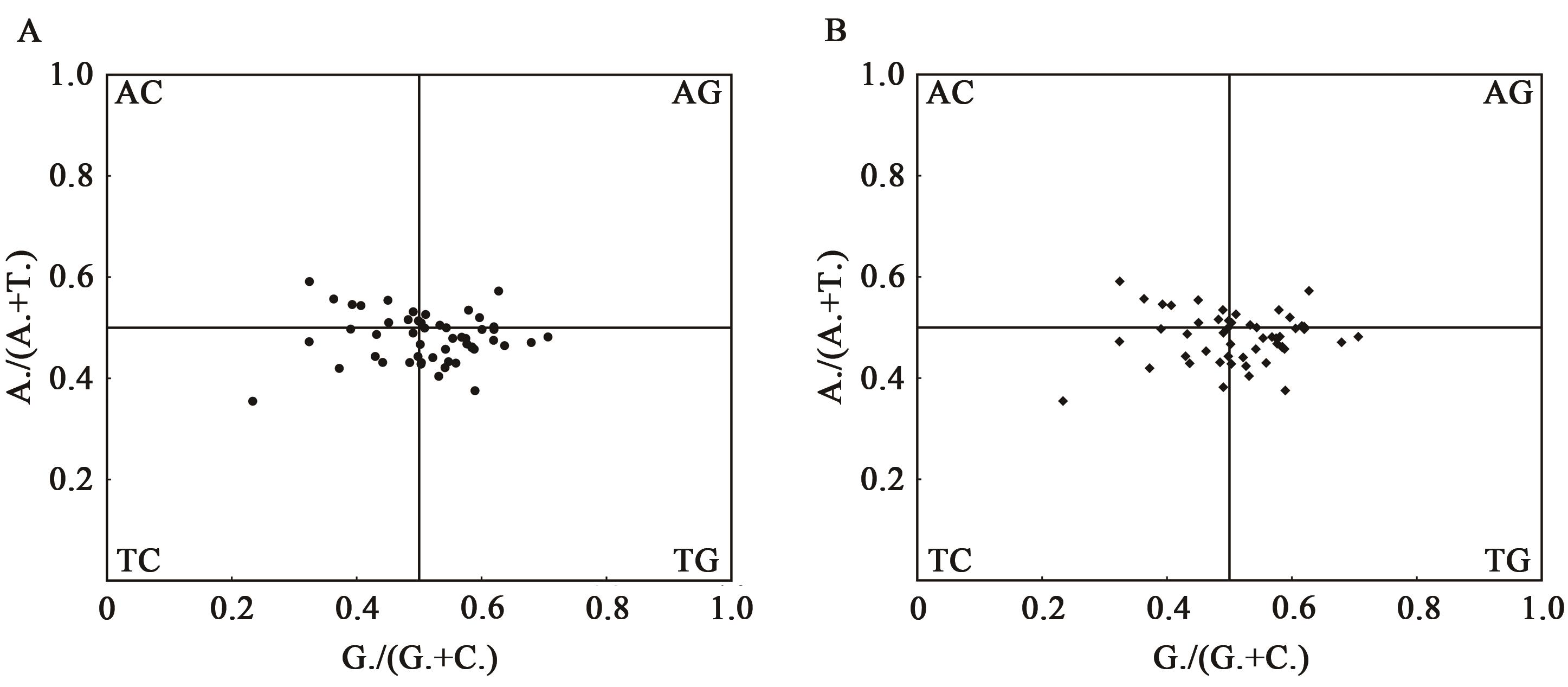
| 物种 | GeneBank ID | GC/% | GC1/% | GC2/% | GC3/% | ENC |
|---|---|---|---|---|---|---|
| 毛叶悬钩子(R. poliophyllus) | PE01833328 | 37.75 | 45.50 | 37.23 | 30.53 | 50.20 |
| 云南悬钩子(R. yunanicus) | HTR285 | 37.77 | 45.43 | 37.39 | 30.49 | 50.20 |
| 高粱泡(R. lambertianus) | NC_056931.1 | 37.78 | 45.44 | 37.42 | 30.48 | 50.17 |
| 蛇泡筋(R. cochinchinensis) | HTR451 | 37.78 | 45.44 | 37.44 | 30.48 | 50.19 |
Table 2 Genome annotation statistics of codon parameter characteristics of chloroplast genome sequences of Rubus plants
| 物种 | GeneBank ID | GC/% | GC1/% | GC2/% | GC3/% | ENC |
|---|---|---|---|---|---|---|
| 毛叶悬钩子(R. poliophyllus) | PE01833328 | 37.75 | 45.50 | 37.23 | 30.53 | 50.20 |
| 云南悬钩子(R. yunanicus) | HTR285 | 37.77 | 45.43 | 37.39 | 30.49 | 50.20 |
| 高粱泡(R. lambertianus) | NC_056931.1 | 37.78 | 45.44 | 37.42 | 30.48 | 50.17 |
| 蛇泡筋(R. cochinchinensis) | HTR451 | 37.78 | 45.44 | 37.44 | 30.48 | 50.19 |
| 氨基酸 | 密码子 | 数目 | RSCU | 氨基酸 | 密码子 | 数目 | RSCU | 氨基酸 | 密码子 | 数目 | RSCU |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 苯丙氨酸 | UUU | 842 | 1.38 | 丝氨酸 | UCU | 473 | 1.54 | AGA | 421 | 1.61 | |
| UUC | 465 | 0.62 | UCC | 309 | 1.01 | AGG | 148 | 0.51 | |||
| 亮氨酸 | UUA | 752 | 2.05 | UCA | 339 | 1.07 | 酪氨酸 | UAU | 673 | 1.57 | |
| UUG | 485 | 1.20 | UCG | 182 | 0.66 | UAC | 170 | 0.36 | |||
| CUU | 534 | 1.33 | AGU | 322 | 1.34 | 终止密码子 | UAA | 28 | 1.50 | ||
| CUC | 160 | 0.33 | AGC | 102 | 0.38 | UAG | 18 | 0.96 | |||
| CUA | 313 | 0.69 | 脯氨酸 | CCU | 349 | 1.40 | UGA | 10 | 0.54 | ||
| CUG | 163 | 0.40 | CCC | 183 | 0.69 | 组氨酸 | CAU | 421 | 1.45 | ||
| 异亮氨酸 | AUU | 952 | 1.52 | CCA | 263 | 1.21 | CAC | 127 | 0.44 | ||
| AUC | 391 | 0.56 | CCG | 142 | 0.62 | 谷氨酰胺 | CAA | 624 | 1.59 | ||
| AUA | 621 | 0.93 | 苏氨酸 | ACU | 431 | 1.49 | CAG | 195 | 0.41 | ||
| 蛋氨酸 | AUG | 539 | 1.00 | ACC | 222 | 0.84 | 天冬酰胺 | AAU | 865 | 1.45 | |
| 结页氨酸 | GUU | 445 | 1.46 | ACA | 349 | 1.21 | AAC | 281 | 0.51 | ||
| GUC | 136 | 0.37 | ACG | 131 | 0.46 | 赖氨酸 | AAA | 940 | 1.57 | ||
| GUA | 456 | 1.56 | 丙氨酸 | GCU | 526 | 1.74 | AAG | 315 | 0.40 | ||
| GUG | 177 | 0.61 | GCC | 191 | 0.64 | 天冬氨酸 | GAU | 767 | 1.53 | ||
| 甘氨酸 | GGU | 485 | 1.32 | GCA | 325 | 1.17 | GAC | 181 | 0.47 | ||
| GGC | 162 | 0.46 | GCG | 135 | 0.45 | 谷氨酸 | GAA | 928 | 1.46 | ||
| GGA | 577 | 1.48 | 精氨酸 | CGU | 289 | 1.59 | GAG | 332 | 0.54 | ||
| GGG | 266 | 0.74 | CGC | 89 | 0.47 | 色氨酸 | UGG | 381 | 0.88 | ||
| 半胱氨酸 | UGU | 194 | 1.43 | CGA | 308 | 1.40 | |||||
| UGC | 61 | 0.42 | CGG | 113 | 0.43 |
Table 3 Relative synonymous codon usage analysis of genes from chloroplast genome in Rubus poliophyllus
| 氨基酸 | 密码子 | 数目 | RSCU | 氨基酸 | 密码子 | 数目 | RSCU | 氨基酸 | 密码子 | 数目 | RSCU |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 苯丙氨酸 | UUU | 842 | 1.38 | 丝氨酸 | UCU | 473 | 1.54 | AGA | 421 | 1.61 | |
| UUC | 465 | 0.62 | UCC | 309 | 1.01 | AGG | 148 | 0.51 | |||
| 亮氨酸 | UUA | 752 | 2.05 | UCA | 339 | 1.07 | 酪氨酸 | UAU | 673 | 1.57 | |
| UUG | 485 | 1.20 | UCG | 182 | 0.66 | UAC | 170 | 0.36 | |||
| CUU | 534 | 1.33 | AGU | 322 | 1.34 | 终止密码子 | UAA | 28 | 1.50 | ||
| CUC | 160 | 0.33 | AGC | 102 | 0.38 | UAG | 18 | 0.96 | |||
| CUA | 313 | 0.69 | 脯氨酸 | CCU | 349 | 1.40 | UGA | 10 | 0.54 | ||
| CUG | 163 | 0.40 | CCC | 183 | 0.69 | 组氨酸 | CAU | 421 | 1.45 | ||
| 异亮氨酸 | AUU | 952 | 1.52 | CCA | 263 | 1.21 | CAC | 127 | 0.44 | ||
| AUC | 391 | 0.56 | CCG | 142 | 0.62 | 谷氨酰胺 | CAA | 624 | 1.59 | ||
| AUA | 621 | 0.93 | 苏氨酸 | ACU | 431 | 1.49 | CAG | 195 | 0.41 | ||
| 蛋氨酸 | AUG | 539 | 1.00 | ACC | 222 | 0.84 | 天冬酰胺 | AAU | 865 | 1.45 | |
| 结页氨酸 | GUU | 445 | 1.46 | ACA | 349 | 1.21 | AAC | 281 | 0.51 | ||
| GUC | 136 | 0.37 | ACG | 131 | 0.46 | 赖氨酸 | AAA | 940 | 1.57 | ||
| GUA | 456 | 1.56 | 丙氨酸 | GCU | 526 | 1.74 | AAG | 315 | 0.40 | ||
| GUG | 177 | 0.61 | GCC | 191 | 0.64 | 天冬氨酸 | GAU | 767 | 1.53 | ||
| 甘氨酸 | GGU | 485 | 1.32 | GCA | 325 | 1.17 | GAC | 181 | 0.47 | ||
| GGC | 162 | 0.46 | GCG | 135 | 0.45 | 谷氨酸 | GAA | 928 | 1.46 | ||
| GGA | 577 | 1.48 | 精氨酸 | CGU | 289 | 1.59 | GAG | 332 | 0.54 | ||
| GGG | 266 | 0.74 | CGC | 89 | 0.47 | 色氨酸 | UGG | 381 | 0.88 | ||
| 半胱氨酸 | UGU | 194 | 1.43 | CGA | 308 | 1.40 | |||||
| UGC | 61 | 0.42 | CGG | 113 | 0.43 |
| 氨基酸 | 密码子 | 数目 | RSCU | 氨基酸 | 密码子 | 数目 | RSCU | 氨基酸 | 密码子 | 数目 | RSCU |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 苯丙氨酸 | UUU | 900 | 1.38 | 丝氨酸 | UCU | 503 | 1.52 | AGA | 445 | 1.61 | |
| UUC | 490 | 0.62 | UCC | 329 | 1.01 | AGG | 162 | 0.53 | |||
| 亮氨酸 | UUA | 813 | 2.07 | UCA | 360 | 1.08 | 酪氨酸 | UAU | 729 | 1.56 | |
| UUG | 524 | 1.21 | UCG | 194 | 0.65 | UAC | 186 | 0.37 | |||
| CUU | 556 | 1.32 | AGU | 363 | 1.35 | 终止密码子 | UAA | 30 | 1.61 | ||
| CUC | 170 | 0.33 | AGC | 116 | 0.39 | UAG | 17 | 0.91 | |||
| CUA | 332 | 0.69 | 脯氨酸 | CCU | 366 | 1.40 | UGA | 9 | 0.48 | ||
| CUG | 170 | 0.38 | CCC | 196 | 0.70 | 组氨酸 | CAU | 452 | 1.44 | ||
| 异亮氨酸 | AUU | 1026 | 1.52 | CCA | 280 | 1.19 | CAC | 140 | 0.45 | ||
| AUC | 412 | 0.55 | CCG | 147 | 0.63 | 谷氨酰胺 | CAA | 666 | 1.59 | ||
| AUA | 661 | 0.93 | 苏氨酸 | ACU | 466 | 1.51 | CAG | 211 | 0.41 | ||
| 蛋氨酸 | AUG | 573 | 1.00 | ACC | 236 | 0.83 | 天冬酰胺 | AAU | 927 | 1.45 | |
| 结页氨酸 | GUU | 477 | 1.48 | ACA | 376 | 1.21 | AAC | 294 | 0.51 | ||
| GUC | 147 | 0.38 | ACG | 137 | 0.45 | 赖氨酸 | AAA | 988 | 1.57 | ||
| GUA | 482 | 1.54 | 丙氨酸 | GCU | 562 | 1.73 | AAG | 328 | 0.39 | ||
| GUG | 187 | 0.60 | GCC | 203 | 0.65 | 天冬氨酸 | GAU | 829 | 1.52 | ||
| 甘氨酸 | GGU | 521 | 1.32 | GCA | 354 | 1.19 | GAC | 198 | 0.48 | ||
| GGC | 177 | 0.46 | GCG | 145 | 0.44 | 谷氨酸 | GAA | 989 | 1.47 | ||
| GGA | 625 | 1.48 | 精氨酸 | CGU | 305 | 1.58 | GAG | 346 | 0.53 | ||
| GGG | 288 | 0.74 | CGC | 96 | 0.48 | 色氨酸 | UGG | 421 | 0.88 | ||
| 半胱氨酸 | UGU | 208 | 1.43 | CGA | 330 | 1.40 | |||||
| UGC | 67 | 0.42 | CGG | 119 | 0.41 |
Table 4 Relative synonymous codon usage analysis of genes from chloroplast genome in Rbubs yunanicus
| 氨基酸 | 密码子 | 数目 | RSCU | 氨基酸 | 密码子 | 数目 | RSCU | 氨基酸 | 密码子 | 数目 | RSCU |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 苯丙氨酸 | UUU | 900 | 1.38 | 丝氨酸 | UCU | 503 | 1.52 | AGA | 445 | 1.61 | |
| UUC | 490 | 0.62 | UCC | 329 | 1.01 | AGG | 162 | 0.53 | |||
| 亮氨酸 | UUA | 813 | 2.07 | UCA | 360 | 1.08 | 酪氨酸 | UAU | 729 | 1.56 | |
| UUG | 524 | 1.21 | UCG | 194 | 0.65 | UAC | 186 | 0.37 | |||
| CUU | 556 | 1.32 | AGU | 363 | 1.35 | 终止密码子 | UAA | 30 | 1.61 | ||
| CUC | 170 | 0.33 | AGC | 116 | 0.39 | UAG | 17 | 0.91 | |||
| CUA | 332 | 0.69 | 脯氨酸 | CCU | 366 | 1.40 | UGA | 9 | 0.48 | ||
| CUG | 170 | 0.38 | CCC | 196 | 0.70 | 组氨酸 | CAU | 452 | 1.44 | ||
| 异亮氨酸 | AUU | 1026 | 1.52 | CCA | 280 | 1.19 | CAC | 140 | 0.45 | ||
| AUC | 412 | 0.55 | CCG | 147 | 0.63 | 谷氨酰胺 | CAA | 666 | 1.59 | ||
| AUA | 661 | 0.93 | 苏氨酸 | ACU | 466 | 1.51 | CAG | 211 | 0.41 | ||
| 蛋氨酸 | AUG | 573 | 1.00 | ACC | 236 | 0.83 | 天冬酰胺 | AAU | 927 | 1.45 | |
| 结页氨酸 | GUU | 477 | 1.48 | ACA | 376 | 1.21 | AAC | 294 | 0.51 | ||
| GUC | 147 | 0.38 | ACG | 137 | 0.45 | 赖氨酸 | AAA | 988 | 1.57 | ||
| GUA | 482 | 1.54 | 丙氨酸 | GCU | 562 | 1.73 | AAG | 328 | 0.39 | ||
| GUG | 187 | 0.60 | GCC | 203 | 0.65 | 天冬氨酸 | GAU | 829 | 1.52 | ||
| 甘氨酸 | GGU | 521 | 1.32 | GCA | 354 | 1.19 | GAC | 198 | 0.48 | ||
| GGC | 177 | 0.46 | GCG | 145 | 0.44 | 谷氨酸 | GAA | 989 | 1.47 | ||
| GGA | 625 | 1.48 | 精氨酸 | CGU | 305 | 1.58 | GAG | 346 | 0.53 | ||
| GGG | 288 | 0.74 | CGC | 96 | 0.48 | 色氨酸 | UGG | 421 | 0.88 | ||
| 半胱氨酸 | UGU | 208 | 1.43 | CGA | 330 | 1.40 | |||||
| UGC | 67 | 0.42 | CGG | 119 | 0.41 |
| 1 | LU L D, BUFFORD D E, eds. Rubus L.[M]//WU Z Y, PETER H R, HONG D Y. Flora of China ( 9). China: Science Press Beijing, 2003:195-285. |
| 2 | 张玉杰,乔娣,马菁霞,等.悬钩子属2中国新记录种[J].西北植物学报,2017,37(4):805-808. |
| ZHANG Y J, QIAO D, MA J X, et al.. Two newly recorded species of Rubus(Rosaceae) from China[J]. Acta Bot. Boreali Occidentalia Sin., 2017, 37(4): 805-808. | |
| 3 | 良良,勤勤,布日额,等.悬钩子木品种整理及其混伪品的DNA条形码鉴定研究[J].中药材,2021,44(9):2069-2073. |
| LIANG L, QIN Q, BU R E, et al.. Study on variety arrangement of Ramulus rubi and DNA barcoding identification of its adulterants[J]. J. Chin. Med. Mater., 2021, 44(9): 2069-2073. | |
| 4 | 俞德浚,陆玲娣.中国植物志[M].第37卷.北京:科学出版社,1985. |
| 5 | 李维林,贺善安.悬钩子属部分类群的分类订正[J].植物研究,2001,21(3):346-349. |
| LI W L, HE S A. Taxonomic revision on several taxa inthe genus Rubus (Rosaceae)[J]. Bull. Bot. Res., 2001, 21(3): 346-349. | |
| 6 | 万静.48种悬钩子属植物表型性状数量分类研究[D].雅安:四川农业大学,2010. |
| 7 | 段娟.西南地区17种(33份)不同来源的野生悬钩子属植物种间遗传多样性的RAPD分析[D].雅安:四川农业大学,2007. |
| 8 | 蒲伟军,谭冰兰,贺丹晨,等.利用重测序技术开发高粱InDel分子标记[J].生物技术进展,2023,13(5):730-741. |
| PU W J, TAN B L, HE D C, et al.. Development of InDel molecular markers in Sorghum using re-sequencing technology[J]. Curr. Biotechnol., 2023, 13(5): 730-741. | |
| 9 | 张丽,王小蓉,王燕,等.DNA序列在悬钩子属植物分子系统学研究中的应用进展[J].西北植物学报,2014,34(2):423-430. |
| ZHANG L, WANG X R, WANG Y, et al.. Research progress of molecular phylogenetic analyses based on DNA sequence data in Rubus L.(Rosaceae)[J]. Acta Bot. Boreali Occidentalia Sin., 2014, 34(2): 423-430. | |
| 10 | DANIELL H, LIN C S, YU M, et al.. Chloroplast genomes: diversity, evolution, and applications in genetic engineering[J/OL]. Genome Biol., 2016, 17(1): 134[2023-11-24]. . |
| 11 | NGUYEN V B, LINH GIANG V N, WAMINAL N E, et al.. Comprehensive comparative analysis of chloroplast genomes from seven Panax species and development of an authentication system based on species-unique single nucleotide polymorphism markers[J]. J. Ginseng Res., 2020, 44(1): 135-144. |
| 12 | MORDEN C W, GARDNER D E, WENIGER D A. Phylogeny and biogeography of Pacific Rubus subgenus idaeobatus (Rosaceae) species: investigating the origin of the endemic Hawaiian raspberry R. macraei [J]. Pac. Sci., 2003, 57(2): 181-197. |
| 13 | IMANISHI H, NAKAHARA K, TSUYUZAKI H. Genetic relationships among native and introduced rubus species in Japan based on rbcl sequence[J]. Acta Hortic., 2008(769): 195-199. |
| 14 | YU J, FU J, FANG Y, et al.. Complete chloroplast genomes of Rubus species (Rosaceae) and comparative analysis within the genus[J/OL]. BMC Genom., 2022, 23(1): 32[2023-11-24]. . |
| 15 | WU L, NIE L, XU Z, et al.. Comparative and phylogenetic analysis of the complete chloroplast genomes of three Paeonia section moutan species (Paeoniaceae)[J/OL]. Front. Genet., 2020, 11: 980[2023-11-24]. . |
| 16 | WANG W, CHEN S, ZHANG X. Whole-genome comparison reveals divergent IR borders and mutation hotspots in chloroplast genomes of herbaceous bamboos (bambusoideae: olyreae)[J/OL]. Molecules, 2018, 23(7): 1537[2023-11-24]. . |
| 17 | 李连星,彭劲谕,王大玮,等.长爪栘[木衣]叶绿体基因组特征系统发育及密码子偏好性分析[J].生物工程学报,2022,38(1):328-342. |
| LI L X, PENG J Y, WANG D W, et al.. Chloroplast genome phylogeny and codon preference of Docynia longiunguis [J]. Chin. J. Biotechnol., 2022, 38(1): 328-342. | |
| 18 | ASAF S, WAQAS M, KHAN A L, et al.. The complete chloroplast genome of wild rice (Oryza minuta) and its comparison to related species[J/OL]. Front. Plant Sci., 2017, 8: 304[2023-11-24]. . |
| 19 | RONO P C, DONG X, YANG J X, et al.. Initial complete chloroplast genomes of Alchemilla (Rosaceae): comparative analysis and phylogenetic relationships[J/OL]. Front. Genet., 2020, 11: 560368[2023-11-24]. . |
| 20 | 赵儒楠,褚晓洁,刘维,等.鹅耳枥属树种叶绿体基因组结构及变异分析[J].南京林业大学学报(自然科学版),2021,45(2):25-34. |
| ZHAO R N, CHU X J, LIU W, et al.. Structure and variation analyses of chloroplast genomes in Carpinus [J]. J. Nanjing For. Univ., 2021, 45(2): 25-34. | |
| 21 | YANG J, CHIANG Y C, HSU T W, et al.. Characterization and comparative analysis among plastome sequences of eight endemic Rubus (Rosaceae) species in Taiwan[J/OL]. Sci. Rep., 2021, 11(1): 1152[2023-11-24]. . |
| 22 | 赵奉彬.中国杨属物种DNA条形码及分子系统学初步研究[D].北京:北京林业大学,2016. |
| 23 | 吴田泽,赵小惠,胡心怡,等.中药材DNA条形码鉴定技术应用进展[J].中医药导报,2019,25(16):125-130. |
| WU T Z, ZHAO X H, HU X Y, et al.. Application progress on DNA barcoding technology of traditional Chinese medicine[J]. Guid. J. Tradit. Chin. Med. Pharm., 2019, 25(16): 125-130. |
| [1] | Hao LIU, Xiang LI. Research Progress on Pathogenic Mechanisms and Treatment Strategy of Coronary Artery Disease [J]. Current Biotechnology, 2025, 15(2): 254-262. |
| [2] | Xu ZHU, Yingnan ZHANG, Jinfa MA, Lihong SHI. The Role of Ptbp1 in T Cell Acute Lymphoblastic Leukemia Mice [J]. Current Biotechnology, 2025, 15(2): 341-348. |
| [3] | Xin QI, Xinran LI, Yaning GUO, Dan WANG, Kai LI, Qiong WU, Liang LI. Comparison of Endogenous Genes in Maize Based on Digital PCR [J]. Current Biotechnology, 2025, 15(1): 78-85. |
| [4] | Guang HU, Zhi WANG, Wei FU, Yuting SHI, Shanshan CHEN, Liang LUO, Shuang WEI. Establishment of Detection Method Based on TaqMan Real-time Fluorescence Quantitative PCR Technology for OsWx-edited Rice [J]. Current Biotechnology, 2025, 15(1): 86-92. |
| [5] | Yin ZHANG, Junwei JIA, Lan BAI, Yifan CHEN, Nguyen CONGTHANH, Beibei LYU, Aihu PAN. Analysis of Method Validation of Transgenic Testing Standards [J]. Current Biotechnology, 2024, 14(1): 120-124. |
| [6] | Yonghong ZHANG, Yanyi LI, Weiting ZHANG. Progress on Integration Site Analysis Technology [J]. Current Biotechnology, 2024, 14(1): 66-71. |
| [7] | Dan YU, Yunlong MA, Fang WAN, Jianqiang WU. Advances on Research and Application of mRNA Vaccines [J]. Current Biotechnology, 2023, 13(4): 492-498. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||