Current Biotechnology ›› 2025, Vol. 15 ›› Issue (3): 418-425.DOI: 10.19586/j.2095-2341.2024.0206
• Reviews • Previous Articles Next Articles
The Research Progress on the Relationship Between Autism and the Three-dimensional Structure of Chromosomes
Haiming GAO( ), Yuanyuan ZHANG, Rong HE(
), Yuanyuan ZHANG, Rong HE( )
)
- Department of Clinical Genetics,Shengjing Hospital,China Medical University,Shenyang 110004,China
-
Received:2024-12-24Accepted:2025-02-27Online:2025-05-25Published:2025-07-01 -
Contact:Rong HE
自闭症与染色质三维结构关系的研究进展
- 中国医科大学附属盛京医院临床遗传科,沈阳 110004
-
通讯作者:何蓉 -
作者简介:高海明 E-mail: gaohm@sj-hospital.org; -
基金资助:辽宁省民生科技计划项目(2021JH2/10300121)
CLC Number:
Cite this article
Haiming GAO, Yuanyuan ZHANG, Rong HE. The Research Progress on the Relationship Between Autism and the Three-dimensional Structure of Chromosomes[J]. Current Biotechnology, 2025, 15(3): 418-425.
高海明, 张媛媛, 何蓉. 自闭症与染色质三维结构关系的研究进展[J]. 生物技术进展, 2025, 15(3): 418-425.
share this article
| 分类 | 子类 | 关键机制 | 参考文献 |
|---|---|---|---|
| 遗传因素 | DNVs | 神经发育相关基因功能破坏 | [ |
| 罕见SNVs/CNVs | 单核苷酸/小片段变异(52%)、染色体结构变异(46%)直接破坏关键基因或调控元件 | [ | |
| 多基因风险评分 | 常见SNPs叠加效应 | [ | |
| 父母生育年龄 | 父系年龄增长导致生殖细胞DNVs增加 | [ | |
| 性别差异 | 女性需更高遗传负荷;X染色体失活和性激素可能增强耐受性 | [ | |
| 表观遗传因素 | 非编码区结构变异 | 破坏启动子/增强子与远端基因的染色质三维互作,干扰时空特异性表达 | [ |
| 环境因素 | 孕期暴露 | 炎症、氧化应激等 | [ |
Table 1 Classification of factors affecting autism
| 分类 | 子类 | 关键机制 | 参考文献 |
|---|---|---|---|
| 遗传因素 | DNVs | 神经发育相关基因功能破坏 | [ |
| 罕见SNVs/CNVs | 单核苷酸/小片段变异(52%)、染色体结构变异(46%)直接破坏关键基因或调控元件 | [ | |
| 多基因风险评分 | 常见SNPs叠加效应 | [ | |
| 父母生育年龄 | 父系年龄增长导致生殖细胞DNVs增加 | [ | |
| 性别差异 | 女性需更高遗传负荷;X染色体失活和性激素可能增强耐受性 | [ | |
| 表观遗传因素 | 非编码区结构变异 | 破坏启动子/增强子与远端基因的染色质三维互作,干扰时空特异性表达 | [ |
| 环境因素 | 孕期暴露 | 炎症、氧化应激等 | [ |
| 机制类型 | 具体机制 | 病理影响 | 参考文献 |
|---|---|---|---|
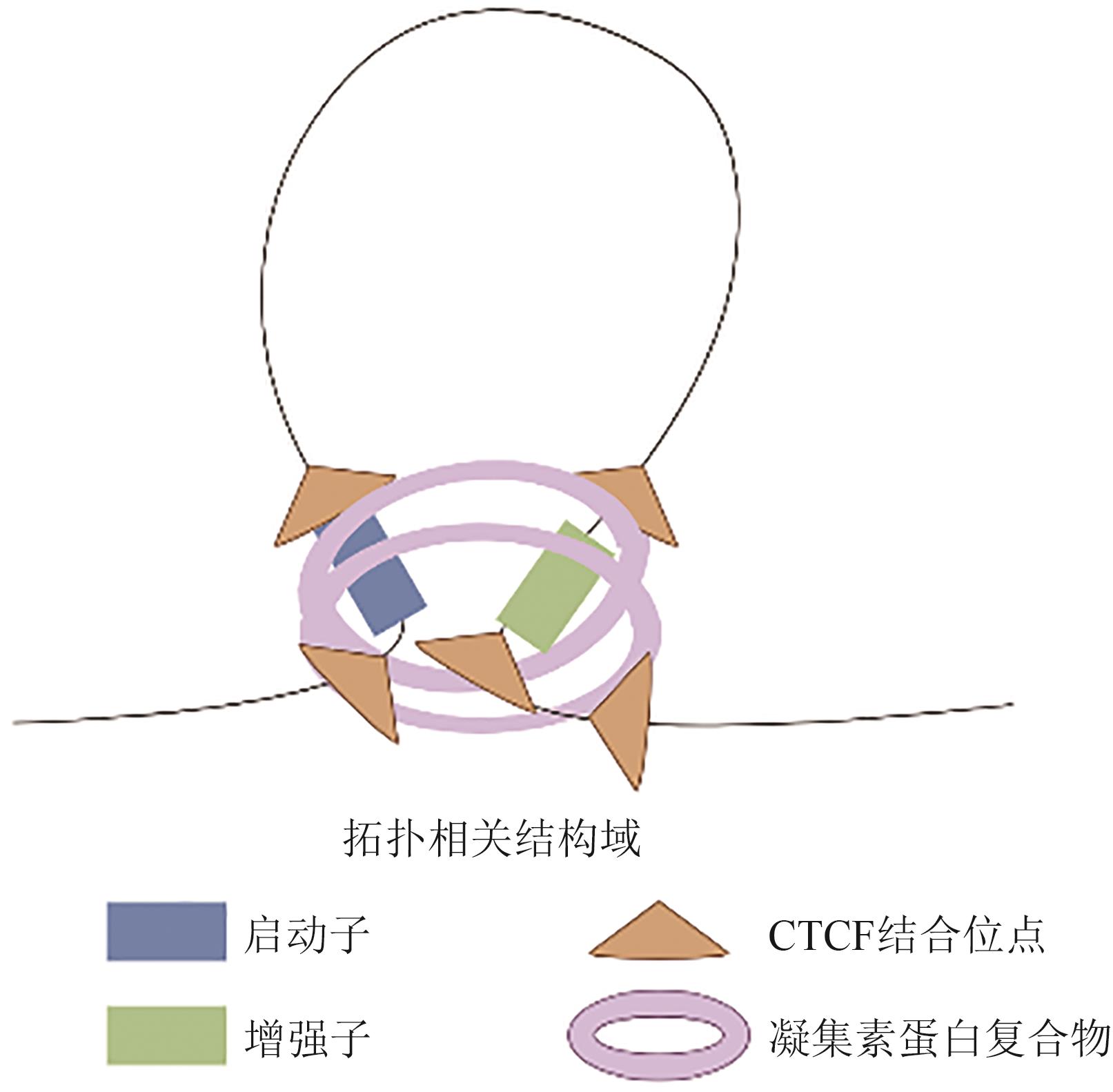
| TAD边界功能异常 | TAD边界表观遗传标记异常或结构破坏,导致增强子-启动子异位互作 | 神经发育关键基因异常表达,影响神经前体细胞增殖与分化 | [ |
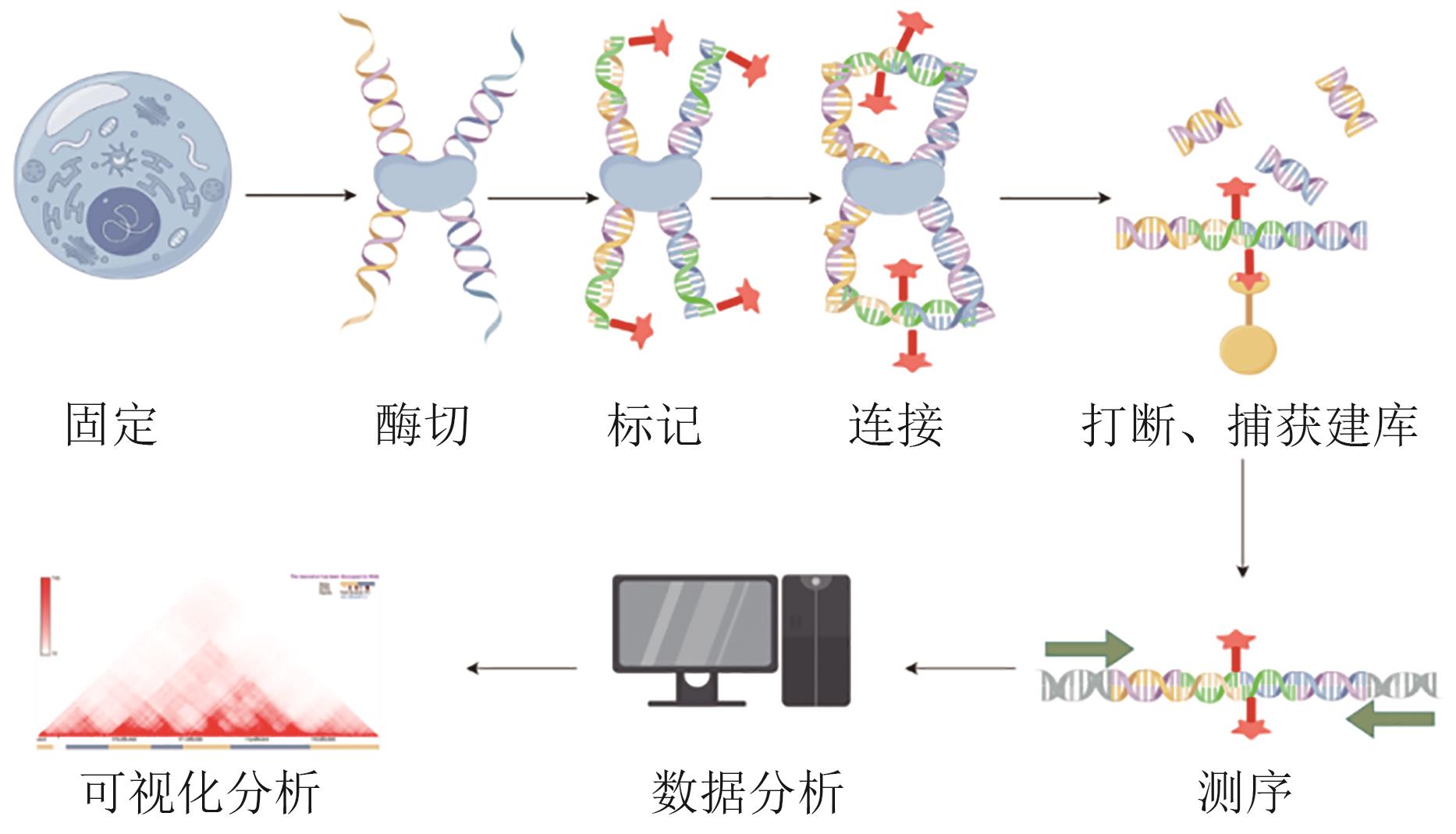
| CTCF蛋白功能失调 | CTCF绝缘边界功能丧失或染色质可及性维持异常,破坏三维基因组稳定性 | 基因异位激活或抑制,多能性基因表达失衡,影响神经分化 | [ |
| 三维互作网络紊乱 | 增强子-启动子长距离或跨染色体互作模式改变,破坏时空特异性转录调控 | 神经发育关键期转录程序失调,引发突触功能及剪接异常 | [ |
| 基因组结构变异 | TADs融合(增强子劫持)或边界缺失(增强子释放-重定向)导致的调控网络重构 | 神经发育相关基因(如皮层形态发生基因)表达异常 | [ |
| 相分离调控失衡 | 转录因子介导的相分离异常,导致染色质动态重组与细胞类型特异性染色质环形成障碍 | 神经细胞命运决定异常,重编程过程受阻 | [ |
| 拓扑结构早发性改变 | 亚核区室化重构、TAD连接强度异常等三维结构变化早于转录异常 | 可作为ASD早期诊断标志,提示神经发育调控网络初始紊乱 | [ |
Table 2 Pathological mechanisms of chromatin three-dimensional structure affecting the occurrence and development of autism
| 机制类型 | 具体机制 | 病理影响 | 参考文献 |
|---|---|---|---|
| TAD边界功能异常 | TAD边界表观遗传标记异常或结构破坏,导致增强子-启动子异位互作 | 神经发育关键基因异常表达,影响神经前体细胞增殖与分化 | [ |
| CTCF蛋白功能失调 | CTCF绝缘边界功能丧失或染色质可及性维持异常,破坏三维基因组稳定性 | 基因异位激活或抑制,多能性基因表达失衡,影响神经分化 | [ |
| 三维互作网络紊乱 | 增强子-启动子长距离或跨染色体互作模式改变,破坏时空特异性转录调控 | 神经发育关键期转录程序失调,引发突触功能及剪接异常 | [ |
| 基因组结构变异 | TADs融合(增强子劫持)或边界缺失(增强子释放-重定向)导致的调控网络重构 | 神经发育相关基因(如皮层形态发生基因)表达异常 | [ |
| 相分离调控失衡 | 转录因子介导的相分离异常,导致染色质动态重组与细胞类型特异性染色质环形成障碍 | 神经细胞命运决定异常,重编程过程受阻 | [ |
| 拓扑结构早发性改变 | 亚核区室化重构、TAD连接强度异常等三维结构变化早于转录异常 | 可作为ASD早期诊断标志,提示神经发育调控网络初始紊乱 | [ |
| 1 | GESCHWIND D H, FLINT J. Genetics and genomics of psychiatric disease[J]. Science, 2015, 349(6255): 1489-1494. |
| 2 | ANTAKI D, GUEVARA J, MAIHOFER A X, et al.. A phenotypic spectrum of autism is attributable to the combined effects of rare variants, polygenic risk and sex[J]. Nat. Genet., 2022, 54(9): 1284-1292. |
| 3 | NAKAMURA T, UEDA J, MIZUNO S, et al.. Topologically associating domains define the impact of de novo promoter variants on autism spectrum disorder risk[J/OL]. Cell Genom., 2024, 4(2): 100488[2025-04-09]. . |
| 4 | CARBALLO-PACORET P, CARRACEDO A, RODRIGUEZ-FONTENLA C. Unraveling the three-dimensional (3D) genome architecture in neurodevelopmental disorders (NDDs)[J]. Neurogenetics, 2024, 25(4): 293-305. |
| 5 | MALACHOWSKI T, CHANDRADOSS K R, BOYA R, et al.. Spatially coordinated heterochromatinization of long synaptic genes in fragile X syndrome[J]. Cell, 2023, 186(26): 5840-5858. |
| 6 | SANDIN S, LICHTENSTEIN P, KUJA-HALKOLA R, et al.. The heritability of autism spectrum disorder[J]. JAMA, 2017, 318(12): 1182-1184. |
| 7 | KAAIJ L J T, MOHN F, VAN DER WEIDE R H, et al.. The ChAHP complex counteracts chromatin looping at CTCF sites that emerged from SINE expansions in mouse[J]. Cell, 2019, 178(6): 1437-1451. |
| 8 | KIM I B, LEE T, LEE J, et al.. Non-coding de novo mutations in chromatin interactions are implicated in autism spectrum disorder[J]. Mol. Psychiatr., 2022, 27(11): 4680-4694. |
| 9 | HONYBUN E, COCKLE E, MALPAS C B, et al.. Neurodevelopmental and functional outcomes following in utero exposure to antiseizure medication: a systematic review[J/OL]. Neurology, 2024, 102(8): e209175[2025-04-09]. . |
| 10 | RUZZO E K, PÉREZ-CANO L, JUNG J Y, et al.. Inherited and de novo genetic risk for autism impacts shared networks[J]. Cell, 2019, 178(4): 850-866. |
| 11 | COHEN S, GABEL H W, HEMBERG M, et al.. Genome-wide activity-dependent MeCP2 phosphorylation regulates nervous system development and function[J]. Neuron, 2011, 72(1): 72-85. |
| 12 | BERNIER R, GOLZIO C, XIONG B, et al.. Disruptive CHD8 mutations define a subtype of autism early in development[J]. Cell, 2014, 158(2): 263-276. |
| 13 | CIRNIGLIARO M, CHANG T S, ARTEAGA S A, et al.. The contributions of rare inherited and polygenic risk to ASD in multiplex families[J/OL]. Proc. Natl. Acad. Sci. USA, 2023, 120(31): e2215632120[2025-04-09]. . |
| 14 | GROTZINGER A D, MALLARD T T, AKINGBUWA W A, et al.. Genetic architecture of 11 major psychiatric disorders at biobehavioral, functional genomic and molecular genetic levels of analysis[J]. Nat. Genet., 2022, 54(5): 548-559. |
| 15 | AN J Y, LIN K, ZHU L, et al.. Genome-wide de novo risk score implicates promoter variation in autism spectrum disorder[J/OL]. Science, 2018, 362(6420): eaat6576[2025-04-09]. . |
| 16 | TROST B, THIRUVAHINDRAPURAM B, CHAN A J S, et al.. Genomic architecture of autism from comprehensive whole-genome sequence annotation[J]. Cell, 2022, 185(23): 4409-4427. |
| 17 | FELICIANO P, ZHOU X, WANG T, et al.. eP121: integrating de novo and inherited variants in over 42607 autism cases identifies variants in new moderate risk genes[J/OL]. Genet. Med., 2022, 24(3): S76[2025-04-09]. . |
| 18 | TURNER T N, COE B P, DICKEL D E, et al.. Genomic patterns of de novo mutation in simplex autism[J]. Cell, 2017, 171(3): 710-722. |
| 19 | DU Z, ZHENG H, HUANG B, et al.. Allelic reprogramming of 3D chromatin architecture during early mammalian development[J]. Nature, 2017, 547(7662): 232-235. |
| 20 | ZHONG H, ZHANG J, LU Y, et al.. 3D genome perspective on cell fate determination, regenerationorgan, and diseases[J/OL]. Cell Prolif., 2023, 56(5): e13482[2025-04-09]. . |
| 21 | FUDENBERG G, IMAKAEV M, LU C, et al.. Formation of chromosomal domains by loop extrusion[J]. Cell Rep., 2016, 15(9): 2038-2049. |
| 22 | TOROSIN N S, ANAND A, GOLLA T R, et al.. 3D genome evolution and reorganization in the Drosophila melanogaster species group[J/OL]. PLoS Genet., 2020, 16(12): e1009229[2025-04-09]. . |
| 23 | ZAGIROVA D, KONONKOVA A, VAULIN N, et al.. From compartments to loops: understanding the unique chromatin organization in neuronal cells[J/OL]. Epigenet. Chromatin, 2024, 17(1): 18[2025-04-09]. . |
| 24 | PHILLIPS-CREMINS J E, SAURIA M E G, SANYAL A, et al.. Architectural protein subclasses shape 3D organization of genomes during lineage commitment[J]. Cell, 2013, 153(6): 1281-1295. |
| 25 | ZUIN J, DIXON J R, VAN DER REIJDEN M I J A, et al.. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells[J]. Proc. Natl. Acad. Sci. USA, 2014, 111(3): 996-1001. |
| 26 | SZABO Q, BANTIGNIES F, CAVALLI G. Principles of genome folding into topologically associating domains[J/OL]. Sci. Adv., 2019, 5(4): eaaw1668[2025-04-09]. . |
| 27 | KREFTING J, ANDRADE-NAVARRO M A, IBN-SALEM J. Evolutionary stability of topologically associating domains is associated with conserved gene regulation[J/OL]. BMC Biol., 2018, 16(1): 87[2025-04-09]. . |
| 28 | MCARTHUR E, CAPRA J A. Topologically associating domain boundaries that are stable across diverse cell types are evolutionarily constrained and enriched for heritability[J]. Am. J. Hum. Genet., 2021, 108(2): 269-283. |
| 29 | ZHANG D, HUANG P, SHARMA M, et al.. Alteration of genome folding via contact domain boundary insertion[J]. Nat. Genet., 2020, 52(10): 1076-1087. |
| 30 | NARENDRA V, ROCHA P P, AN D, et al.. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation[J]. Science, 2015, 347(6225): 1017-1021. |
| 31 | DE RUBEIS S, HE X, GOLDBERG A P, et al.. Synaptic, transcriptional and chromatin genes disrupted in autism[J]. Nature, 2014, 515(7526): 209-215. |
| 32 | LU H, YU D, HANSEN A S, et al.. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase Ⅱ[J]. Nature, 2018, 558(7709): 318-323. |
| 33 | SONG Y, LIANG Z, ZHANG J, et al.. CTCF functions as an insulator for somatic genes and a chromatin remodeler for pluripotency genes during reprogramming[J/OL]. Cell Rep., 2022, 39(1): 110626[2025-04-09]. . |
| 34 | ANDREY G, MUNDLOS S. The three-dimensional genome: regulating gene expression during pluripotency and development[J]. Development, 2017, 144(20): 3646-3658. |
| 35 | OH S, SHAO J, MITRA J, et al.. Enhancer release and retargeting activates disease-susceptibility genes[J]. Nature, 2021, 595(7869): 735-740. |
| 36 | MAKOWSKI C, VAN DER MEER D, DONG W, et al.. Discovery of genomic loci of the human cerebral cortex using genetically informed brain atlases[J]. Science, 2022, 375(6580): 522-528. |
| 37 | WANG J, YU H, MA Q, et al.. Phase separation of OCT4 controls TAD reorganization to promote cell fate transitions[J]. Cell Stem Cell, 2021, 28(10): 1868-1883. |
| 38 | KRIJGER P H L, DI STEFANO B, DE WIT E, et al.. Cell-of-origin-specific 3D genome structure acquired during somatic cell reprogramming[J]. Cell Stem Cell, 2016, 18(5): 597-610. |
| 39 | BEAGAN J A, PASTUZYN E D, FERNANDEZ L R, et al.. Three-dimensional genome restructuring across timescales of activity-induced neuronal gene expression[J]. Nat. Neurosci., 2020, 23(6): 707-717. |
| 40 | WILFERT A B, TURNER T N, MURALI S C, et al.. Recent ultra-rare inherited variants implicate new autism candidate risk genes[J]. Nat. Genet., 2021, 53(8): 1125-1134. |
| 41 | ANANIA C, ACEMEL R D, JEDAMZICK J, et al.. In vivo dissection of a clustered-CTCF domain boundary reveals developmental principles of regulatory insulation[J]. Nat. Genet., 2022, 54(7): 1026-1036. |
| 42 | HUYNH L, HORMOZDIARI F. TAD fusion score: discovery and ranking the contribution of deletions to genome structure[J/OL]. Genome Biol., 2019, 20(1): 60[2025-04-09]. . |
| 43 | JERKOVIC I, CAVALLI G. Understanding 3D genome organization by multidisciplinary methods[J]. Nat. Rev. Mol. Cell Biol., 2021, 22(8): 511-528. |
| 44 | MOHANTA T K, MISHRA A K, AL-HARRASI A. The 3D genome: from structure to function[J/OL]. Int. J. Mol. Sci., 2021, 22(21): 11585[2025-04-09]. . |
| 45 | LIEBERMAN-AIDEN E, VAN BERKUM N L, WILLIAMS L, et al.. Comprehensive mapping of long-range interactions reveals folding principles of the human genome[J]. Science, 2009, 326(5950): 289-293. |
| 46 | MOTA-GÓMEZ I, LUPIÁÑEZ D G. A (3D-nuclear) space odyssey: making sense of Hi-C maps[J/OL]. Genes, 2019, 10(6): 415[2025-04-09]. . |
| 47 | RAO S S P, HUNTLEY M H, DURAND N C, et al.. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping[J]. Cell, 2014, 159(7): 1665-1680. |
| 48 | SIKORSKA N, SEXTON T. Defining functionally relevant spatial chromatin domains: it is a TAD complicated[J]. J. Mol. Biol., 2020, 432(3): 653-664. |
| 49 | ZHANG S, HE Y, LIU H, et al.. regBase: whole genome base-wise aggregation and functional prediction for human non-coding regulatory variants[J/OL]. Nucleic Acids Res., 2019, 47(21): e134[2025-04-09]. . |
| 50 | WANG Z, ZHAO G, LI B, et al.. Performance comparison of computational methods for the prediction of the function and pathogenicity of non-coding variants[J]. Genom. Proteom. Bioinform., 2023, 21(3): 649-661. |
| 51 | HE Z, LIU L, BELLOY M E, et al.. Ghost knockoff inference empowers identification of putative causal variants in genome-wide association studies[J/OL]. Nat. Commun., 2022, 13(1): 7209[2025-04-09]. . |
| 52 | HE Z, LIU L, WANG C, et al.. Identification of putative causal loci in whole-genome sequencing data via knockoff statistics[J/OL]. Nat. Commun., 2021, 12(1): 3152[2025-04-09]. . |
| [1] | Jingjing FANG, Kunlun HUANG, Tao TONG. Research Progress on Experimental Models of Autism Spectrum Disorders [J]. Current Biotechnology, 2023, 13(4): 509-523. |
| [2] | YUAN Qifeng, YAO Baozhen*. Progress of Abnormal Glutamate-glutamine Cycle in Autism Spectrum Disorders [J]. Curr. Biotech., 2021, 11(2): 170-175. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||