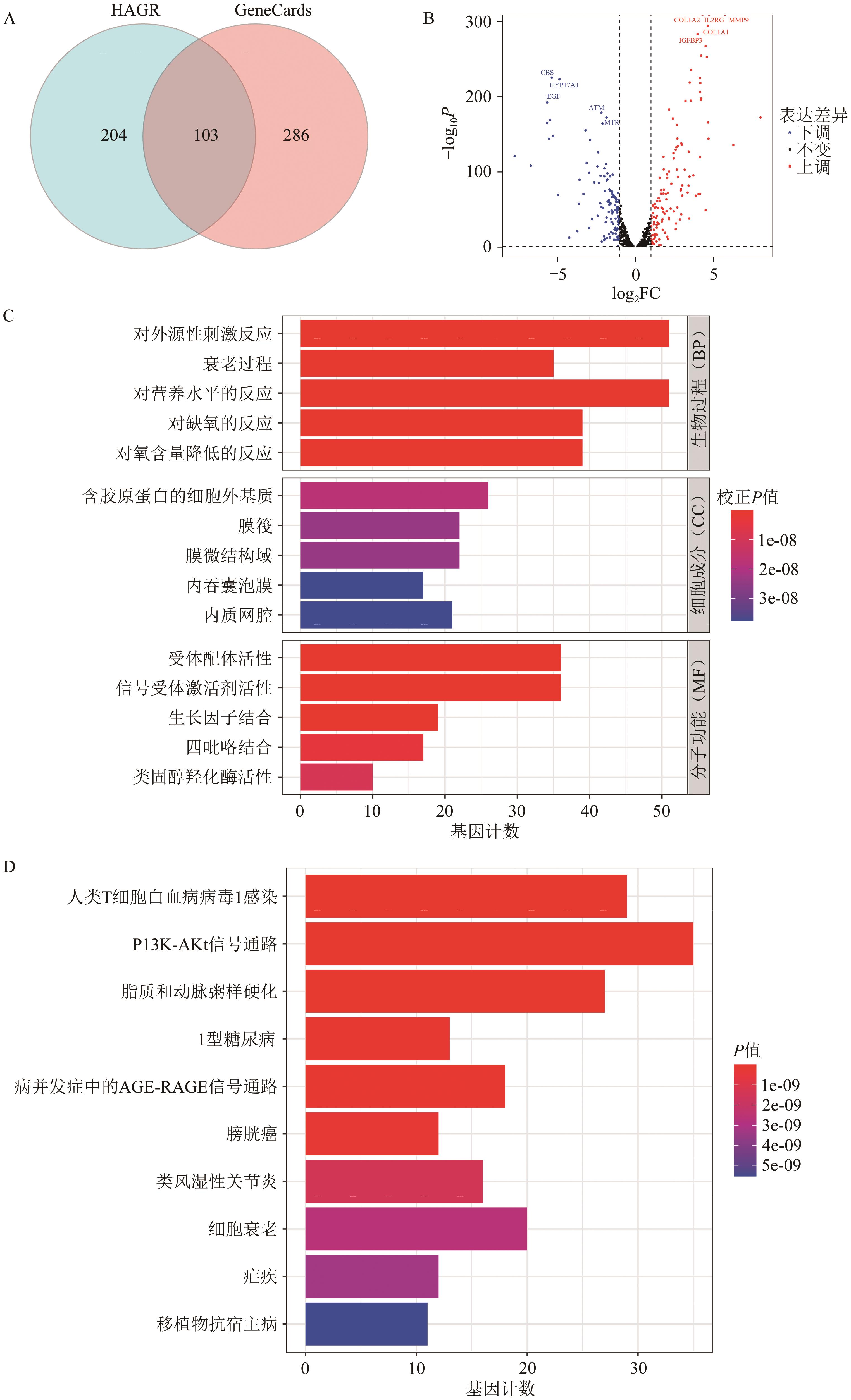
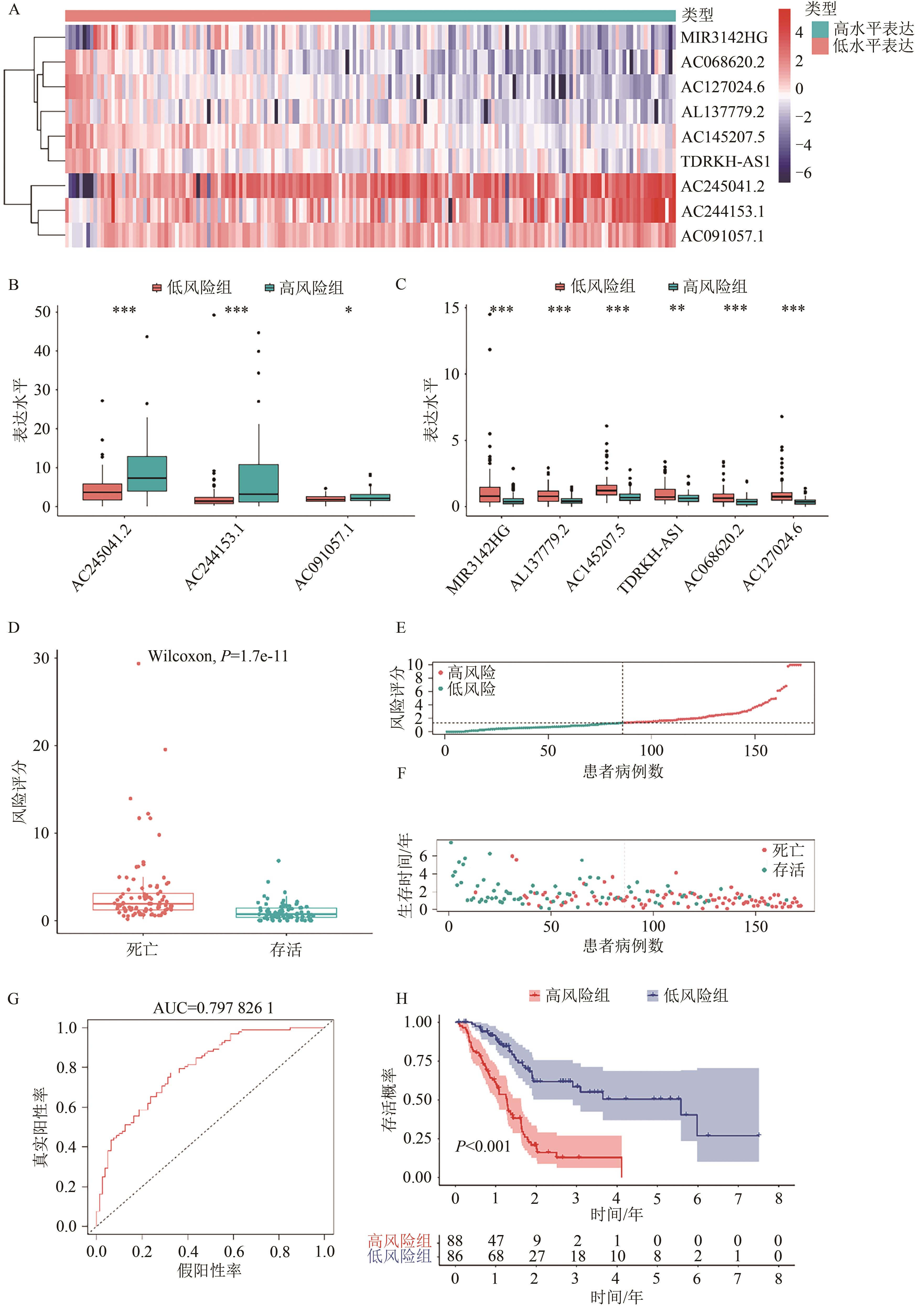
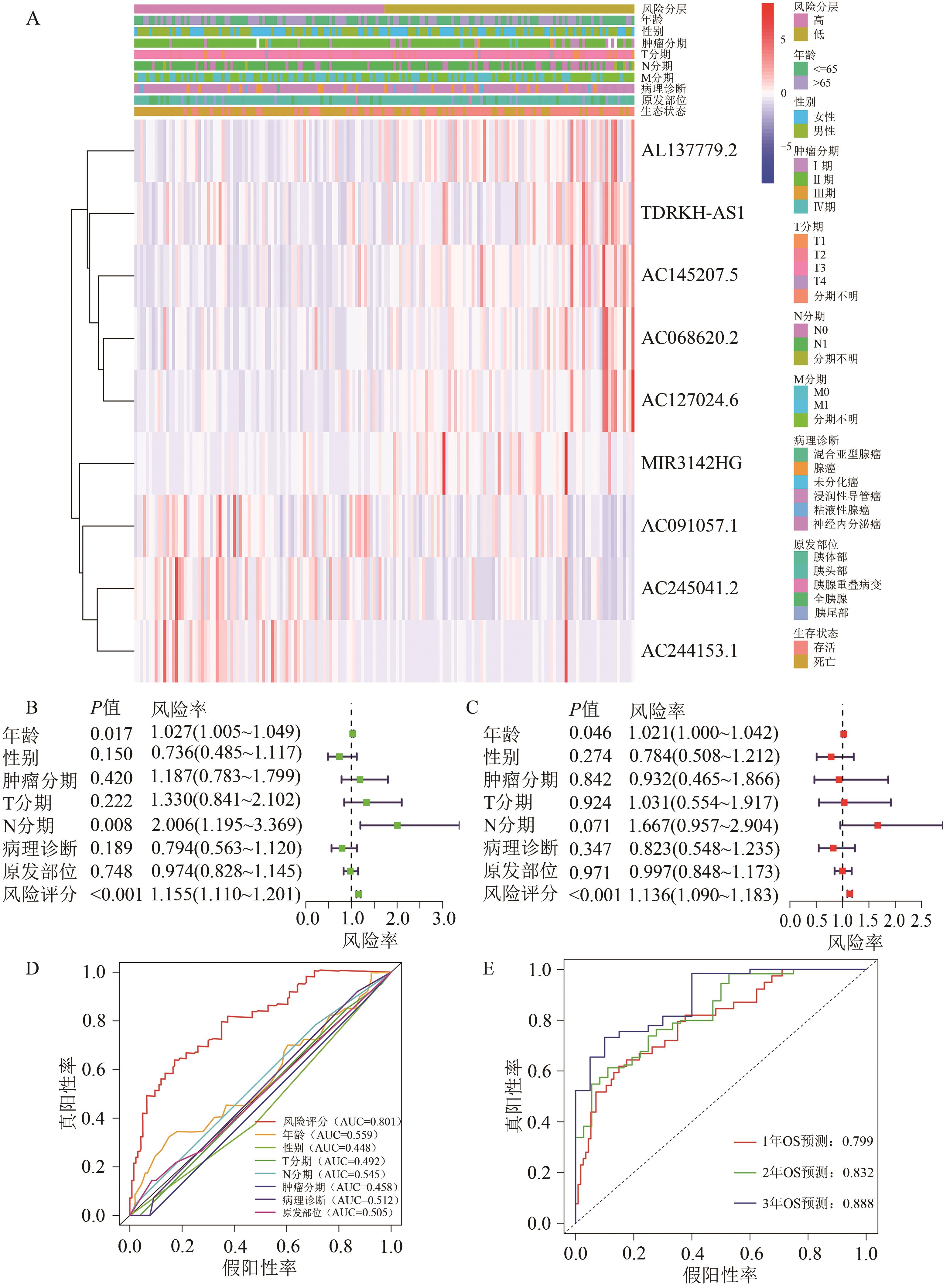
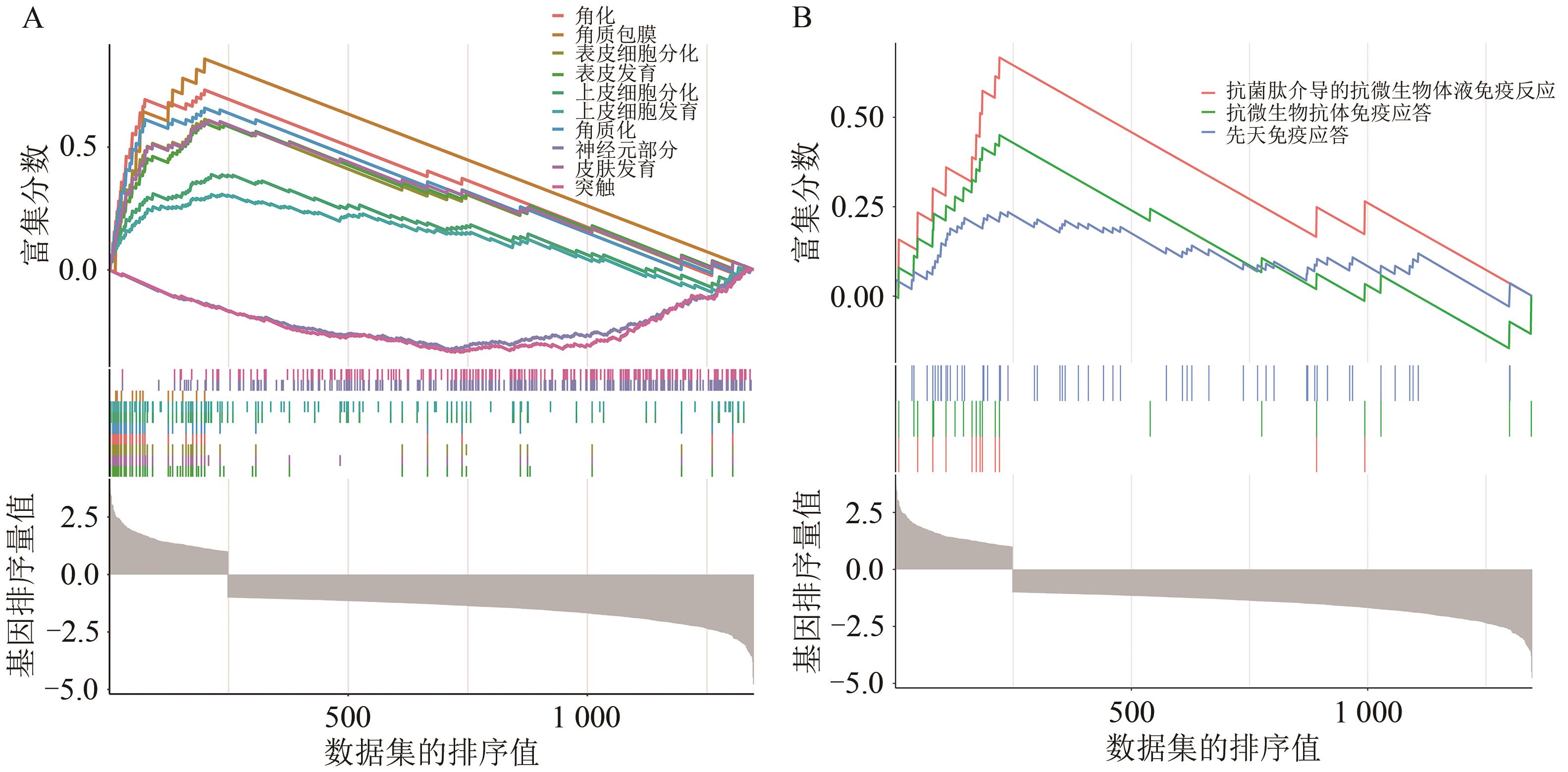
Current Biotechnology ›› 2025, Vol. 15 ›› Issue (1): 158-169.DOI: 10.19586/j.2095-2341.2024.0027
• Articles • Previous Articles Next Articles
Pancreatic Cancer Risk Stratification and Prognostic Prediction Model Based on Aging-related Gene Characteristics
Xinxiong LI1( ), Weixuan HONG2, Tianshun FENG3, Junwei FANG2, Hu ZHAO2, Chunhong XIAO2, Meiping WANG2(
), Weixuan HONG2, Tianshun FENG3, Junwei FANG2, Hu ZHAO2, Chunhong XIAO2, Meiping WANG2( )
)
- 1.Department of General Surgery,Dongfang Hospital,Xiamen University,Fuzhou 350025,China
2.Department of General Surgery,the 900th Hospital of Joint Logistics Support Force,Fuzhou 350025,China
3.Department of Neurosurgery,Dongfang Hospital,Xiamen University,Fuzhou 350025,China
-
Received:2024-02-20Accepted:2024-07-02Online:2025-01-25Published:2025-03-07 -
Contact:Meiping WANG
基于衰老相关基因特征的胰腺癌风险分层及预后预测模型
李新雄1( ), 洪伟煊2, 冯添顺3, 房俊伟2, 赵虎2, 肖春红2, 王梅平2(
), 洪伟煊2, 冯添顺3, 房俊伟2, 赵虎2, 肖春红2, 王梅平2( )
)
- 1.厦门大学附属东方医院普通外科,福州 350025
2.联勤保障部队第九〇〇医院普通外科,福州 350025
3.厦门大学附属东方医院神经外科,福州 350025
-
通讯作者:王梅平 -
作者简介:李新雄 E-mail: xx1129552947@163.com; -
基金资助:福建省自然科学基金项目(2021J011262);联勤保障部队第九〇〇医院青年自主创新项目孵化专项(2022QC07)
CLC Number:
Cite this article
Xinxiong LI, Weixuan HONG, Tianshun FENG, Junwei FANG, Hu ZHAO, Chunhong XIAO, Meiping WANG. Pancreatic Cancer Risk Stratification and Prognostic Prediction Model Based on Aging-related Gene Characteristics[J]. Current Biotechnology, 2025, 15(1): 158-169.
李新雄, 洪伟煊, 冯添顺, 房俊伟, 赵虎, 肖春红, 王梅平. 基于衰老相关基因特征的胰腺癌风险分层及预后预测模型[J]. 生物技术进展, 2025, 15(1): 158-169.
share this article
| 1 | HUANG J, LOK V, NGAI C H, et al.. Worldwide burden of, risk factors for, and trends in pancreatic cancer[J]. Gastroenterology, 2021, 160(3): 744-754. |
| 2 | SIEGEL R L, MILLER K D, WAGLE N S, et al.. Cancer statistics, 2023[J]. Cancer J. Clin, 2023, 73(1): 17-48. |
| 3 | CALCINOTTO A, KOHLI J, ZAGATO E, et al.. Cellular senescence: aging, cancer, and injury[J]. Physiol. Rev., 2019, 99(2): 1047-1078. |
| 4 | SAUL D, KOSINSKY R L. Single-cell transcriptomics reveals the expression of aging- and senescence-associated genes in distinct cancer cell populations[J/OL]. Cells, 2021, 10(11): 3126[2024-03-12]. . |
| 5 | ZABRANSKY D J, JAFFEE E M, WEERARATNA A T. Shared genetic and epigenetic changes link aging and cancer[J]. Trends Cell Biol., 2022, 32(4): 338-350. |
| 6 | FANE M, WEERARATNA A T. How the ageing microenvironment influences tumour progression[J]. Nat. Rev. Cancer, 2020, 20(2): 89-106. |
| 7 | QUINN J J, CHANG H Y. Unique features of long non-coding RNA biogenesis and function[J]. Nat. Rev. Genet., 2016, 17(1): 47-62. |
| 8 | STATELLO L, GUO C J, CHEN L L, et al.. Gene regulation by long non-coding RNAs and its biological functions[J]. Nat. Rev. Mol. Cell Biol., 2021, 22(2): 96-118. |
| 9 | HUARTE M. The emerging role of lncRNAs in cancer[J]. Nat. Med., 2015, 21(11): 1253-1261. |
| 10 | BHAN A, SOLEIMANI M, MANDAL S S. Long noncoding RNA and cancer: a new paradigm[J]. Cancer Res., 2017, 77(15): 3965-3981. |
| 11 | ROBLESS E E, HOWARD J A, CASARI I, et al.. Exosomal long non-coding RNAs in the diagnosis and oncogenesis of pancreatic cancer[J]. Cancer Lett., 2021, 501: 55-65. |
| 12 | LI D, HE J, QIAN X, et al.. The involvement of lncRNAs in the development and progression of pancreatic cancer[J]. Cancer Biol. Ther., 2017, 18(12): 927-936. |
| 13 | VIVIAN J, RAO A A, NOTHAFT F A, et al.. Toil enables reproducible, open source, big biomedical data analyses[J]. Nat. Biotechnol., 2017, 35(4): 314-316. |
| 14 | YU G, WANG L G, HAN Y, et al.. clusterProfiler: an R package for comparing biological themes among gene clusters[J]. Omics, 2012, 16(5): 284-287. |
| 15 | BRUNSON J C. Ggalluvial: layered grammar for alluvial plots[J/OL]. J. Open Source Softw., 2020, 5(49): 2017[2024-03-12]. . |
| 16 | NEWMAN A M, LIU C L, GREEN M R, et al.. Robust enumeration of cell subsets from tissue expression profiles[J]. Nat. Meth., 2015, 12(5): 453-457. |
| 17 | BECHT E, GIRALDO N A, LACROIX L, et al.. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression[J/OL]. Genome Biol., 2016, 17(1): 218[2024-03-12]. doi: 10.1186/s13059-016-1070-5 .. |
| 18 | HÄNZELMANN S, CASTELO R, GUINNEY J. GSVA gene set variation analysis for microarray and RNA-seq data[J/OL]. BMC Bioinform., 2013, 14: 7[2024-03-12]. . |
| 19 | LENZO F L, KATO S, PABLA S, et al.. Immune profiling and immunotherapeutic targets in pancreatic cancer[J/OL]. Ann. Transl. Med., 2021, 9(2): 119[2024-03-12]. . |
| 20 | PING H, JIA X, KE H. A novel ferroptosis-related lncRNAs signature predicts clinical prognosis and is associated with immune landscape in pancreatic cancer[J/OL]. Front. Genet., 2022, 13: 786689[2024-03-12]. . |
| 21 | KIRKEGÅRD J, MORTENSEN F V, HANSEN C P, et al.. Waiting time to surgery and pancreatic cancer survival: a nationwide population-based cohort study[J]. Eur. J. Surg. Oncol., 2019, 45(10): 1901-1905. |
| 22 | van der GEEST L G M, van EIJCK C H J, GROOT KOERKAMP B, et al.. Trends in treatment and survival of patients with nonresected, nonmetastatic pancreatic cancer: a population-based study[J]. Cancer Med., 2018, 7(10): 4943-4951. |
| 23 | YUAN J, DUAN F, ZHAI W, et al.. An aging-related gene signature-based model for risk stratification and prognosis prediction in breast cancer[J]. Int. J. Women Health, 2021, 13: 1053-1064. |
| 24 | WANG D, NING H, WU H, et al.. Construction and evaluation of a novel prognostic risk model of aging-related genes in bladder cancer[J]. Curr. Urol., 2023, 17(4): 236-245. |
| 25 | XU Q, CHEN Y. An aging-related gene signature-based model for risk stratification and prognosis prediction in lung adenocarcinoma[J/OL]. Front. Cell Dev. Biol., 2021, 9: 685379[2024-03-12]. . |
| 26 | 刘文增,胡渊,张彩.胰腺癌的肿瘤微环境及其免疫治疗研究进展[J].中国免疫学杂志,2018,34(12):1901-1906. |
| LIU W Z, HU Y, ZHANG C. Research progress in tumor microenvironment and immunotherapy of pancreatic cancer[J]. Chin. J. Immunol., 2018, 34(12): 1901-1906. | |
| 27 | TABIBZADEH S. Signaling pathways and effectors of aging[J]. Front. Biosci. Landmark Ed., 2021, 26(1): 50-96. |
| 28 | FLEMING T H, HUMPERT P M, NAWROTH P P, et al.. Reactive metabolites and AGE/RAGE-mediated cellular dysfunction affect the aging process: a mini-review[J]. Gerontology, 2011, 57(5): 435-443. |
| 29 | WAGHELA B N, VAIDYA F U, RANJAN K, et al.. AGE-RAGE synergy influences programmed cell death signaling to promote cancer[J]. Mol. Cell. Biochem., 2021, 476(2): 585-598. |
| 30 | REN X, CHEN C, LUO Y, et al.. lncRNA-PLACT1 sustains activation of NF-κB pathway through a positive feedback loop with IκBα/E2F1 axis in pancreatic cancer[J/OL]. Mol. Cancer, 2020, 19(1): 35[2024-03-12]. . |
| 31 | LIN J, ZHAI S, ZOU S, et al.. Positive feedback between lncRNA FLVCR1-AS1 and KLF10 may inhibit pancreatic cancer progression via the PTEN/AKT pathway[J/OL]. J. Exp. Clin. Cancer Res., 2021, 40(1): 316[2024-03-12]. . |
| 32 | CHENG Q, OUYANG X, ZHANG R, et al.. Senescence-associated genes and non-coding RNAs function in pancreatic cancer progression[J]. RNA Biol., 2020, 17(11): 1693-1706. |
| 33 | JIANG W, DU Y, ZHANG W, et al.. Construction of a prognostic model based on cuproptosis-related lncRNA signatures in pancreatic cancer[J/OL]. Can. J. Gastroenterol. Hepatol., 2022, 2022: 4661929[2024-03-12]. . |
| 34 | ZHAO K, LI X, SHI Y, et al.. A comprehensive analysis of pyroptosis-related lncRNAs signature associated with prognosis and tumor immune microenvironment of pancreatic adenocarcinoma[J/OL]. Front. Genet., 2022, 13: 899496[2024-03-12]. . |
| 35 | LI J, ZHANG J, TAO S, et al.. Prognostication of pancreatic cancer using the cancer genome atlas based ferroptosis-related long non-coding RNAs[J/OL]. Front. Genet., 2022, 13: 838021[2024-03-12]. . |
| 36 | JIAO Y, ZHOU J, JIN Y, et al.. Long non-coding RNA TDRKH-AS1 promotes colorectal cancer cell proliferation and invasion through the β-catenin activated Wnt signaling pathway[J/OL]. Front. Oncol., 2020, 10: 639[2024-03-12]. . |
| 37 | TAUBE J M, GALON J, SHOLL L M, et al.. Implications of the tumor immune microenvironment for staging and therapeutics[J]. Mod. Pathol., 2018, 31(2): 214-234. |
| 38 | LIN H J, LIU Y, CAROLAND K, et al.. Polarization of cancer-associated macrophages maneuver neoplastic attributes of pancreatic ductal adenocarcinoma[J/OL]. Cancers, 2023, 15(13): 3507[2024-03-12]. . |
| 39 | CHOUEIRY F, TOROK M, SHAKYA R, et al.. CD200 promotes immunosuppression in the pancreatic tumor microenvironment[J/OL]. J. Immunother. Cancer, 2020, 8(1): e000189[2024-03-12]. . |
| 40 | LIU S, ZHANG W, LIU K, et al.. CD160 expression on CD8+ T cells is associated with active effector responses but limited activation potential in pancreatic cancer[J]. Cancer Immunol. Immunother., 2020, 69(5): 789-797. |
| 41 | VONDERHEIDE R H, BAYNE L J. Inflammatory networks and immune surveillance of pancreatic carcinoma[J]. Curr. Opin. Immunol., 2013, 25(2): 200-205. |
| 42 | 陈俊俊,黄浩,刘颖婷,等.胰腺癌免疫微环境组织驻留CD103+CD8+T细胞浸润分布及其临床意义[J].中华肿瘤防治杂志,2022,29(23):1659-1667. |
| CHEN J J, HUANG H, LIU Y T, et al.. Infiltration of tissue-resident CD103+CD8+T cells in the tumor microenvironment of pancreatic cancer and its clinical significance[J]. Chin. J. Cancer Prev. Treat., 2022, 29(23): 1659-1667. | |
| 43 | CHIBAYA L, MURPHY K C, DEMARCO K D, et al.. EZH2 inhibition remodels the inflammatory senescence-associated secretory phenotype to potentiate pancreatic cancer immune surveillance[J]. Nat. Cancer, 2023, 4(6): 872-892. |
| [1] | Ziyi ZHANG-HUANG, Lisha HUANG, Yanqi LI, Chenlu XIONG, Ying YU, Fei XIE. Construction of a Breast Cancer Prognostic Model Based on Nicotine Metabolism Gene Signatures [J]. Current Biotechnology, 2025, 15(4): 735-742. |
| [2] | Xu ZHU, Yingnan ZHANG, Jinfa MA, Lihong SHI. The Role of Ptbp1 in T Cell Acute Lymphoblastic Leukemia Mice [J]. Current Biotechnology, 2025, 15(2): 341-348. |
| [3] | Jinqing ZHONG, Kun HU, Xiaoning GAO, Di FANG, Zehui SU, Siyu ZHANG, Chengwei HUANG. In vivo Study of p,p'-DDT Bioconcentration Kinetics in Chinese Mitten Crab Eriocheir sinensis [J]. Current Biotechnology, 2024, 14(5): 848-856. |
| [4] | Lingyue ZHENG, Jingwei WANG, Jingyuan TONG, Lihong SHI. The Establishment of A Novel Effective Method for the Enrichment of Murine Bone Marrow Erythroblastic Islands [J]. Current Biotechnology, 2024, 14(3): 466-472. |
| [5] | Lili SUN, Yuemaierabola ANWAIER, Fuzhong LIU, Yeerkenbieke BUERLAN, Ye DILINAER, Wenjia GUO. Construction of Prognostic Prediction Model of Breast Cancer Based on Tumor-associated Fibroblast Genes and Analysis of Immune Infiltration [J]. Current Biotechnology, 2024, 14(2): 312-322. |
| [6] | Ting XU, Jiahao SHEN, Kang ZHAO, Lu HUANG, Enhui DONG, Kexin ZENG, Xinwei BIAN, Minghui JI, Qin XU. Bacterial Signature for Prediction of Disease Type Based on Abundance of Ruminococcus [J]. Current Biotechnology, 2024, 14(2): 323-330. |
| [7] | Yuemaierabola ANWAIER, Yeerkenbieke BUERLAN, Lili SUN, Fuzhong LIU, Yeerxiati DILINAER, Wenjia GUO. Prognosis Prediction Model and Drug Sensitivity Analysis of Triple-negative Breast Cancer Based on m5C Related Genes [J]. Current Biotechnology, 2024, 14(1): 149-159. |
| [8] | Xin LIU, Hua LI, Lei QI, Lixia YANG, Yuexin YU, Lanju XU. Therapeutic Effect of Recombinant Collagen Gynecological Gel on Cervicitis in Rats [J]. Current Biotechnology, 2024, 14(1): 42-47. |
| [9] | Jiayuan JIAO, Jiuge XING, Baoshan CHAI. Research Progress on RET Oncogene in Thyroid Carcinoma [J]. Current Biotechnology, 2023, 13(6): 895-899. |
| [10] | Haitao CAO, Jing ZHU, Yunpeng MA, Xinghua CUI. Application of Machine Learning in Phenotypic Prediction of Gut Microbiota [J]. Current Biotechnology, 2023, 13(5): 671-680. |
| [11] | Haitao CAO, Jing ZHU, Haibo ZENG, Yanchen LIU. Research on Feature Selection of Gut Microbiota and Disease Prediction Model Based on Weighted Average [J]. Current Biotechnology, 2023, 13(5): 798-806. |
| [12] | Ali WANG, Jiangdong LIU. Research Progress on the CRISPR/Cas System in Zebrafish [J]. Current Biotechnology, 2023, 13(4): 485-491. |
| [13] | Jingjing FANG, Kunlun HUANG, Tao TONG. Research Progress on Experimental Models of Autism Spectrum Disorders [J]. Current Biotechnology, 2023, 13(4): 509-523. |
| [14] | Yunpeng MA, Jing ZHU, Xinghua CUI. Content Estimating of Microbial Dissolved Organic Carbon Based on Machine Learning [J]. Current Biotechnology, 2023, 13(4): 645-653. |
| [15] | Wei ZHANG, Hongfang WANG, Baohua XU. Overview of the Main Molecular Mechanisms of Biological Aging [J]. Current Biotechnology, 2023, 13(2): 228-233. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||