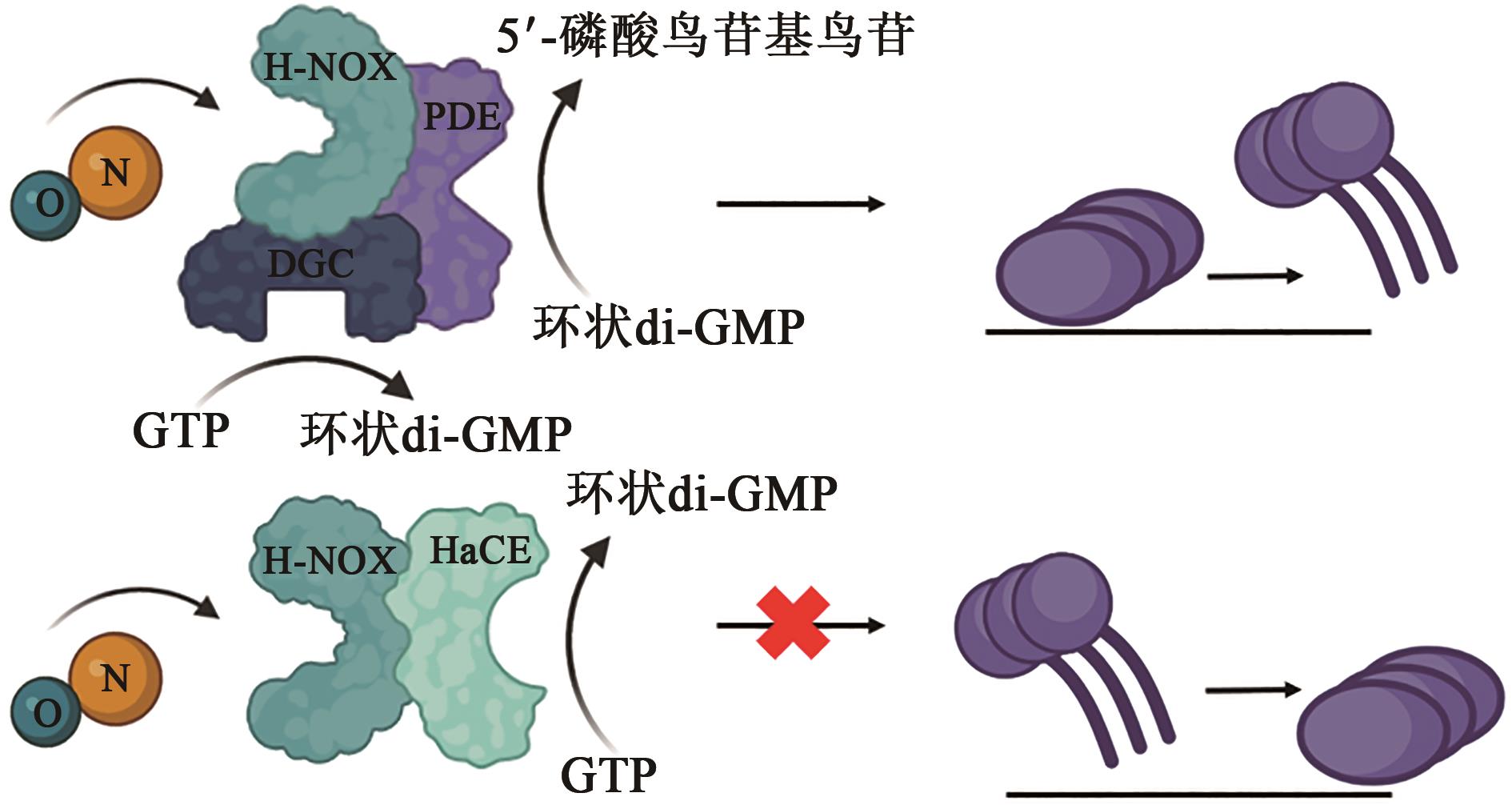
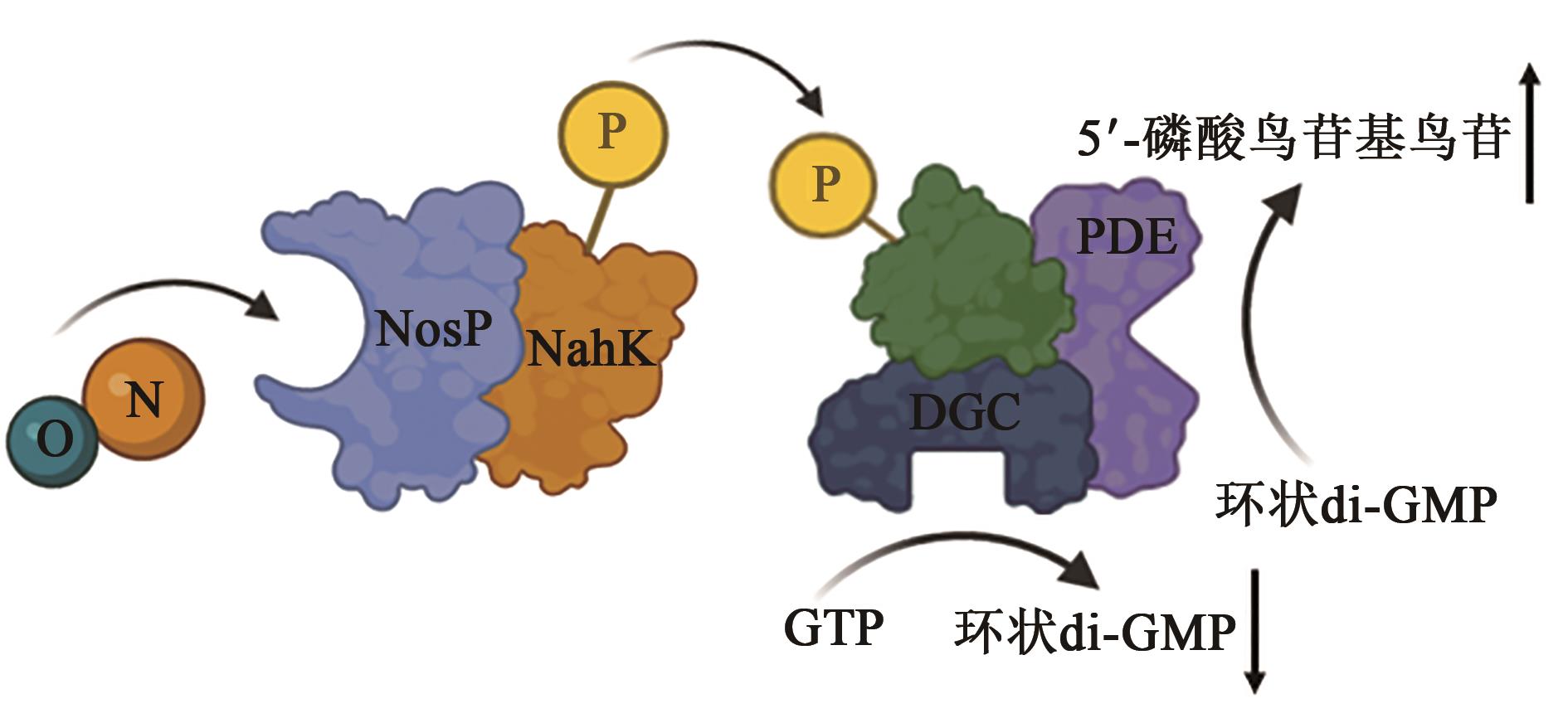
| 1 |
WAREHAM L K, SOUTHAM H M, POOLE R K. Do nitric oxide, carbon monoxide and hydrogen sulfide really qualify as 'gasotransmitters' in bacteria?[J]. Biochem. Soc. Trans., 2018, 46(5): 1107-1118.
|
| 2 |
CUI Q, YANG Y, JI N, et al.. Gaseous signaling molecules and their application in resistant cancer treatment: from invisible to visible[J]. Future Med. Chem., 2019, 11(4): 323-336.
|
| 3 |
DE PAULA T D, SILVA B R, GRANDO M D, et al.. Relaxation induced by the nitric oxide donor and cyclooxygenase inhibitor NCX2121 in renal hypertensive rat aortas[J]. Eur. J. Pharm. Sci. Off. J. Eur. Fed. Pharm. Sci., 2017, 107: 45-53.
|
| 4 |
KASHFI K. Carbon monoxide in cell signaling and potential therapeutics[J/OL]. Biochem. Pharmacol., 2022, 204: 115231[2022-08-19]. .
|
| 5 |
SHAN H, QIU J, CHANG P, et al.. Exogenous hydrogen sulfide offers neuroprotection on intracerebral hemorrhage injury through modulating endogenous H2S metabolism in mice[J/OL]. Front. Cell. Neurosci., 2019, 13: 349[2019-08-07]. .
|
| 6 |
YANG S Q, JIANG L, LAN F, et al.. Inhibited endogenous H2S generation and excessive autophagy in hippocampus contribute to sleep deprivation-induced cognitive impairment[J/OL]. Front. Psychol., 2019, 10: 53[2019-01-24]. .
|
| 7 |
KULKARNI-CHITNIS M, MITCHELL-BUSH L, BELFORD R, et al.. Interaction between hydrogen sulfide, nitric oxide, and carbon monoxide pathways in the bovine isolated retina[J]. AIMS Neurosci., 2019, 6(3): 104-115.
|
| 8 |
AUSMA T, DE KOK L J. Atmospheric H2S: impact on plant functioning[J/OL]. Front. Plant Sci., 2019, 10: 743[2019-06-11]. .
|
| 9 |
LIN C C, YANG C C, HSIAO L D, et al.. Heme oxygenase-1 induction by carbon monoxide releasing molecule-3 suppresses interleukin-1 β-mediated neuroinflammation[J/OL]. Front. Mol. Neurosci., 2017, 10: 387[2017-11-20]. .
|
| 10 |
MENDES S S, MIRANDA V, SARAIVA L M. Hydrogen sulfide and carbon monoxide tolerance in bacteria[J/OL]. Antioxidants, 2021, 10(5): 729[2021-05-05]. .
|
| 11 |
SOUSA E H, LOPES L G, GONZALEZ G, et al.. Drug discovery targeting heme-based sensors and their coupled activities[J]. J. Inorg. Biochem., 2017, 167: 12-20.
|
| 12 |
KRÜGER A, KEPPEL M, SHARMA V, et al.. The diversity of heme sensor systems-heme-responsive transcriptional regulation mediated by transient heme protein interactions[J/OL]. FEMS Microbiol. Rev., 2022, 46(3): fuac002[2022-05-06]. .
|
| 13 |
CORREIA S S, IYENGAR R R, GERMANO P, et al.. The CNS-penetrant soluble guanylate cyclase Stimulator CY6463 reveals its therapeutic potential in neurodegenerative diseases[J/OL]. Front. Pharmacol., 2021, 12: 656561[2021-05-24]. .
|
| 14 |
SUMI M P, TUPTA B, GHOSH A. Nitric oxide trickle drives heme into hemoglobin and muscle myoglobin[J/OL]. Cells, 2022, 11(18): 2838[2022-09-12]. .
|
| 15 |
ROBERTS G P, THORSTEINSSON M V, KERBY R L, et al.. CooA: a heme-containing regulatory protein that serves as a specific sensor of both carbon monoxide and redox state[J]. Prog. Nucl. Acid Res. Mol. Biol., 2001, 67: 35-63.
|
| 16 |
MOHRMANN H, DRAGELJ J, BASERGA F, et al.. The reductive phase of Rhodobacter sphaeroides cytochrome c oxidase disentangled by CO ligation[J/OL]. Phys. Chem. Chem. Phys., 2017,19:32143[2014-11-16]. .
|
| 17 |
NISHINAGA M, SUGIMOTO H, NISHITANI Y, et al.. Heme controls the structural rearrangement of its sensor protein mediating the hemolytic bacterial survival[J/OL]. Commun. Biol., 2021, 4(1): 467[2021-04-13]. .
|
| 18 |
CHILDERS K C, YAO X Q, GIANNAKOULIAS S, et al.. Synergistic mutations in soluble guanylyl cyclase (sGC) reveal a key role for interfacial regions in the sGC activation mechanism[J]. J. Biol. Chem., 2019, 294(48): 18451-18464.
|
| 19 |
KOESLING D, MERGIA E, RUSSWURM M. Physiological functions of NO-sensitive guanylyl cyclase isoforms[J]. Curr. Med. Chem., 2016, 23(24): 2653-2665.
|
| 20 |
DERBYSHIRE E R, MARLETTA M A. Structure and regulation of soluble guanylate cyclase[J]. Annu. Rev. Biochem., 2012, 81: 533-559.
|
| 21 |
MONTFORT W R, WALES J A, WEICHSEL A. Structure and activation of soluble guanylyl cyclase, the nitric oxide sensor[J]. Antioxid. Redox Signal., 2017, 26(3): 107-121.
|
| 22 |
RÜHLE A, ELGERT C, HAHN M G, et al.. Tyrosine 135 of the β1 subunit as binding site of BAY-543: importance of the Y-x-S-x-R motif for binding and activation by sGC activator drugs[J/OL]. Eur. J. Pharmacol., 2020, 881: 173203[2020-05-13]. .
|
| 23 |
KANG Y, LIU R, WU J X, et al.. Structural insights into the mechanism of human soluble guanylate cyclase[J]. Nature, 2019, 574(7777): 206-210.
|
| 24 |
LIU T, SCHROEDER H, POWER G G, et al.. A physiologically relevant role for NO stored in vascular smooth muscle cells: a novel theory of vascular NO signaling[J/OL]. Redox Biol., 2022, 53: 102327[2022-05-09]. .
|
| 25 |
SÖMMER A, BEHRENDS S. Methods to investigate structure and activation dynamics of GC-1/GC-2[J]. Nitric Oxide, 2018, 78: 127-139.
|
| 26 |
BACON B, NISBETT L M, BOON E. Bacterial haemoprotein sensors of NO: H-NOX and NosP[J]. Adv. Microb. Physiol., 2017, 70: 1-36.
|
| 27 |
L-MNISBETT, BOON E M. Nitric oxide regulation of H-NOX signaling pathways in bacteria[J]. Biochemistry, 2016, 55(35): 4873-4884.
|
| 28 |
PELLICENA P, KAROW D S, BOON E M, et al.. Crystal structure of an oxygen-binding heme domain related to soluble guanylate cyclases[J]. Proc. Natl. Acad. Sci. USA, 2004, 101(35): 12854-12859.
|
| 29 |
KHALID R R, SIDDIQI A R, MYLONAS E, et al.. Dynamic characterization of the human heme nitric oxide/oxygen (HNOX) domain under the influence of diatomic gaseous ligands[J/OL]. Int. J. Mol. Sci., 2019, 20(3): 698[2019-02-06]. .
|
| 30 |
CHEN C Y, LEE W, RENHOWE P A, et al.. Solution structures of the Shewanella woodyi H-NOX protein in the presence and absence of soluble guanylyl cyclase stimulator IWP-051[J]. Protein Sci., 2021, 30(2): 448-463.
|
| 31 |
KAROW D S, PAN D, TRAN R, et al.. Spectroscopic characterization of the soluble guanylate cyclase-like heme domains from Vibrio cholerae and Thermoanaerobacter tengcongensis [J]. Biochemistry, 2004, 43(31): 10203-10211.
|
| 32 |
HOSSAIN S, BOON E M. Discovery of a novel nitric oxide binding protein and nitric-oxide-responsive signaling pathway in Pseudomonas aeruginosa [J]. ACS Infect. Dis., 2017, 3(6): 454-461.
|
| 33 |
BACON B A, LIU Y, KINCAID J R, et al.. Spectral characterization of a novel NO sensing protein in bacteria: NosP[J]. Biochemistry, 2018, 57(43): 6187-6200.
|
| 34 |
MA X, SAYED N, BEUVE A, et al.. NO and CO differentially activate soluble guanylyl cyclase via a heme pivot-bend mechanism[J]. EMBO J., 2007, 26(2): 578-588.
|
| 35 |
VOS M H, SALMAN M, LIEBL U. Early processes in heme-based CO-sensing proteins[J/OL]. Front. Mol. Biosci., 2022, 9: 1046412[2022-11-03]. .
|
| 36 |
BORJIGIN M, LI H, LANZ N D, et al.. Structure-based hypothesis on the activation of the CO-sensing transcription factor CooA[J]. Acta Crystallogr. Sect. D Biol. Crystallogr., 2007, 63(3): 282-287.
|
| 37 |
HINES J P, DENT M R, STEVENS D J, et al.. Site-directed spin label electron paramagnetic resonance spectroscopy as a probe of conformational dynamics in the Fe(Ⅲ) "locked-off" state of the CO-sensing transcription factor CooA[J]. Protein Sci. Publ. Protein Soc., 2018, 27(9): 1670-1679.
|
| 38 |
ROBERTS G P, KERBY R L, YOUN H, et al.. CooA, a paradigm for gas sensing regulatory proteins[J]. J. Inorg. Biochem., 2005, 99(1): 280-292.
|
| 39 |
TRIPATHI S, POULOS T L. Testing the N-terminal velcro model of CooA carbon monoxide activation[J]. Biochemistry, 2018, 57(21): 3059-3064.
|
| 40 |
KERBY R L, ROBERTS G P. Burkholderia xenovorans RcoM(Bx)-1, a transcriptional regulator system for sensing low and persistent levels of carbon monoxide[J]. J. Bacteriol., 2012, 194(21): 5803-5816.
|
| 41 |
KERBY R L, YOUN H, ROBERTS G P. RcoM: a new single-component transcriptional regulator of CO metabolism in bacteria[J]. J. Bacteriol., 2008, 190(9): 3336-3343.
|
| 42 |
BOWMAN H E, DENT M R, BURSTYN J N. Met(104) is the CO-replaceable ligand at Fe(Ⅱ) heme in the CO-sensing transcription factor BxRcoM-1[J]. J. Biol. Inorg. Chem., 2016, 21(4): 559-569.
|
| 43 |
SALMAN M, VILLAMIL F C, RAMODIHARILAFY R, et al.. Interaction of the full-length heme-based CO sensor protein RcoM-2 with ligands[J]. Biochemistry, 2019, 58(39): 4028-4034.
|
| 44 |
GIMÉNEZ-MASCARELL P, MAJTAN T, OYENARTE I, et al.. Crystal structure of cystathionine β-synthase from honeybee Apis mellifera [J]. J. Struct. Biol., 2018, 202(1): 82-93.
|
| 45 |
PEY A L, MARTÍNEZ-CRUZ L A, KRAUS J P, et al.. Oligomeric status of human cystathionine beta-synthase modulates AdoMet binding[J]. FEBS Lett., 2016, 590(24): 4461-4471.
|
| 46 |
ANASHKIN V A, BAYKOV A A, LAHTI R. Enzymes regulated via cystathionine β-synthase domains[J]. Biochem. Biokhimiia, 2017, 82(10): 1079-1087.
|
| 47 |
BHATT A, MUKHOPADHYAYA A, ALI M E. α-Helix in cystathionine β-synthase enzyme acts as an electron reservoir[J]. J. Phys. Chem. B, 2022, 126(26): 4754-4760.
|
| 48 |
SUEMATSU M, NAKAMURA T, TOKUMOTO Y, et al.. CO-CBS-H2 S axis: from vascular mediator to cancer regulator[J]. Microcirculation, 2016, 23(3): 183-190.
|
| 49 |
KABE Y, YAMAMOTO T, KAJIMURA M, et al.. Cystathionine β-synthase and PGRMC1 as CO sensors[J]. Free. Radic. Biol. Med., 2016, 99: 333-344.
|
| 50 |
TUPTA B, STUEHR E, SUMI M P, et al.. GAPDH is involved in the heme-maturation of myoglobin and hemoglobin[J/OL]. FASEB J., 2022, 36(2): e22099[2022-12-31]. .
|
| 51 |
KAWAHARA B, FAULL K F, JANZEN C, et al.. Carbon monoxide inhibits cytochrome P450 enzymes CYP3A4/2C8 in human breast cancer cells, increasing sensitivity to paclitaxel[J]. J. Med. Chem., 2021, 64(12): 8437-8446.
|
| 52 |
YANG P M, HUANG Y T, ZHANG Y Q, et al.. Carbon monoxide releasing molecule induces endothelial nitric oxide synthase activation through a calcium and phosphatidylinositol 3-kinase/Akt mechanism[J]. Vasc. Pharmacol., 2016, 87: 209-218.
|
| 53 |
WANG J, LI X, CHANG J W, et al.. Enzymological and structural characterization of Arabidopsis thaliana heme oxygenase-1[J]. FEBS Open Bio., 2022, 12(9): 1677-1687.
|
| 54 |
LIU S, XIA S, YUE D, et al.. The bonding nature of Fe-CO complexes in heme proteins[J]. Inorg. Chem., 2022, 61(44): 17494-17504.
|
| 55 |
FARIS P, NEGRI S, FARIS D, et al.. Hydrogen sulfide (H2S): as a potent modulator and therapeutic prodrug in cancer[J]. Curr. Med. Chem., 2023, 30(40): 4506-45032.
|
| 56 |
ZOU S, SHIMIZU T, YAMAMOTO M, et al.. Hydrogen sulfide-induced relaxation of the bladder is attenuated in spontaneously hypertensive rats[J]. Int. Urol. Nephrol., 2019, 51(9): 1507-1515.
|
| 57 |
JIA J, WANG Z, ZHANG M, et al.. SQR mediates therapeutic effects of H2S by targeting mitochondrial electron transport to induce mitochondrial uncoupling[J/OL]. Sci. Adv., 2020, 6(35): eaaz5752[2020-08-26]. .
|
| 58 |
GHEIBI S, JEDDI S, KASHFI K, et al.. Regulation of vascular tone homeostasis by NO and H2S: implications in hypertension[J]. Biochem. Pharmacol., 2018, 149: 42-59.
|
| 59 |
BARROW K, WANG Y, YU R, et al.. H2S protects from oxidative stress-driven ACE2 expression and cardiac aging[J]. Mol. Cell. Biochem., 2022, 477(5): 1393-1403.
|
| 60 |
JIANG J L, TIAN Y, LI L, et al.. H2S alleviates salinity stress in cucumber by maintaining the Na+/K+ balance and regulating H2S metabolism and oxidative stress response[J/OL]. Front. Plant Sci., 2019, 10: 678[2019-05-28]. .
|
| 61 |
CORPAS F J, PALMA J M. H2S signaling in plants and applications in agriculture[J]. J. Adv. Res., 2020, 24: 131-137.
|
| 62 |
LIU F, ZHANG X, CAI B, et al.. Physiological response and transcription profiling analysis reveal the role of glutathione in H2S-induced chilling stress tolerance of cucumber seedlings[J/OL]. Plant Sci. Int. J. Exp. Plant Biol., 2020, 291: 110363[2019-11-29]. .
|
| 63 |
SHIMIZU T, MASUDA S. Persulphide-responsive transcriptional regulation and metabolism in bacteria[J]. J. Biochem., 2020, 167(2): 125-132.
|
| 64 |
JIN Z, ZHAO P, GONG W, et al.. Fe-porphyrin: a redox-related biosensor of hydrogen molecule[J]. Nano Res., 2023, 16(2): 2020-2025.
|
| 65 |
YOU Y, ZHU Y X, JIANG J, et al.. Water-enabled H2 generation from hydrogenated silicon nanosheets for efficient anti-inflammation[J]. J. Am. Chem. Soc., 2022, 144(31): 14195-14206.
|
| 66 |
TONG J, ZHANG Y, YU P, et al.. Protective effect of hydrogen gas on mouse hind limb ischemia-reperfusion injury[J]. J. Surg. Res., 2021, 266: 148-159.
|
| 67 |
王濛,仪杨,孙梦婷,等.富氢水和富氢生理盐水生物医学研究进展——动物实验[J].生物技术进展, 2022, 12(3): 332-343.
|
| 68 |
马雪梅,张鑫,谢飞,等.氢气生物学作用的生物酶基础[J].生物技术进展, 2020,10(1):15-22.
|
| 69 |
ZHANG Y, XU J, YANG H. Hydrogen: an endogenous regulator of liver homeostasis[J/OL]. Front. Pharmacol., 2020, 11: 877[2020-06-11]. .
|
 ), Xuemei MA(
), Xuemei MA( )
)
 ), 马雪梅(
), 马雪梅( )
)