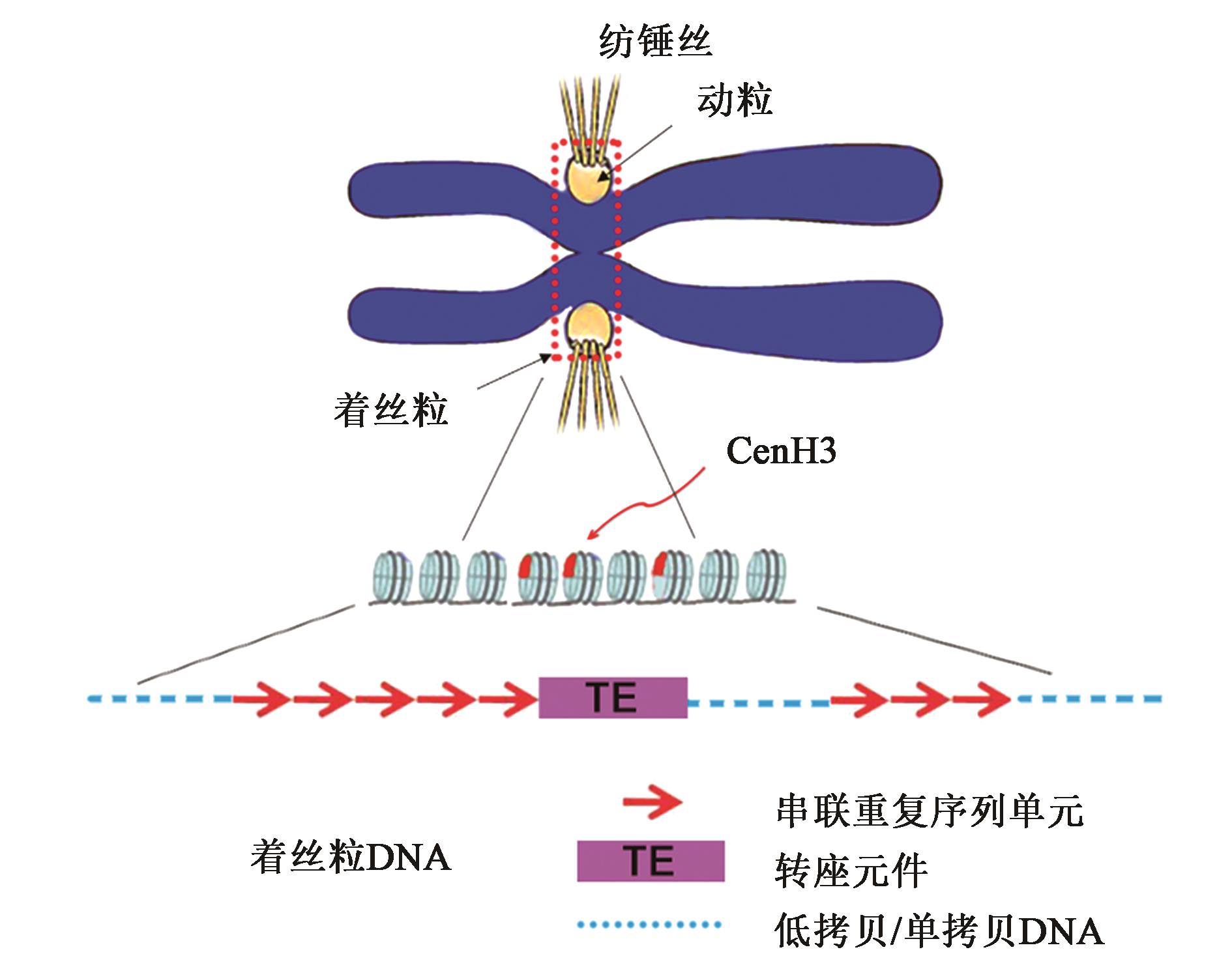
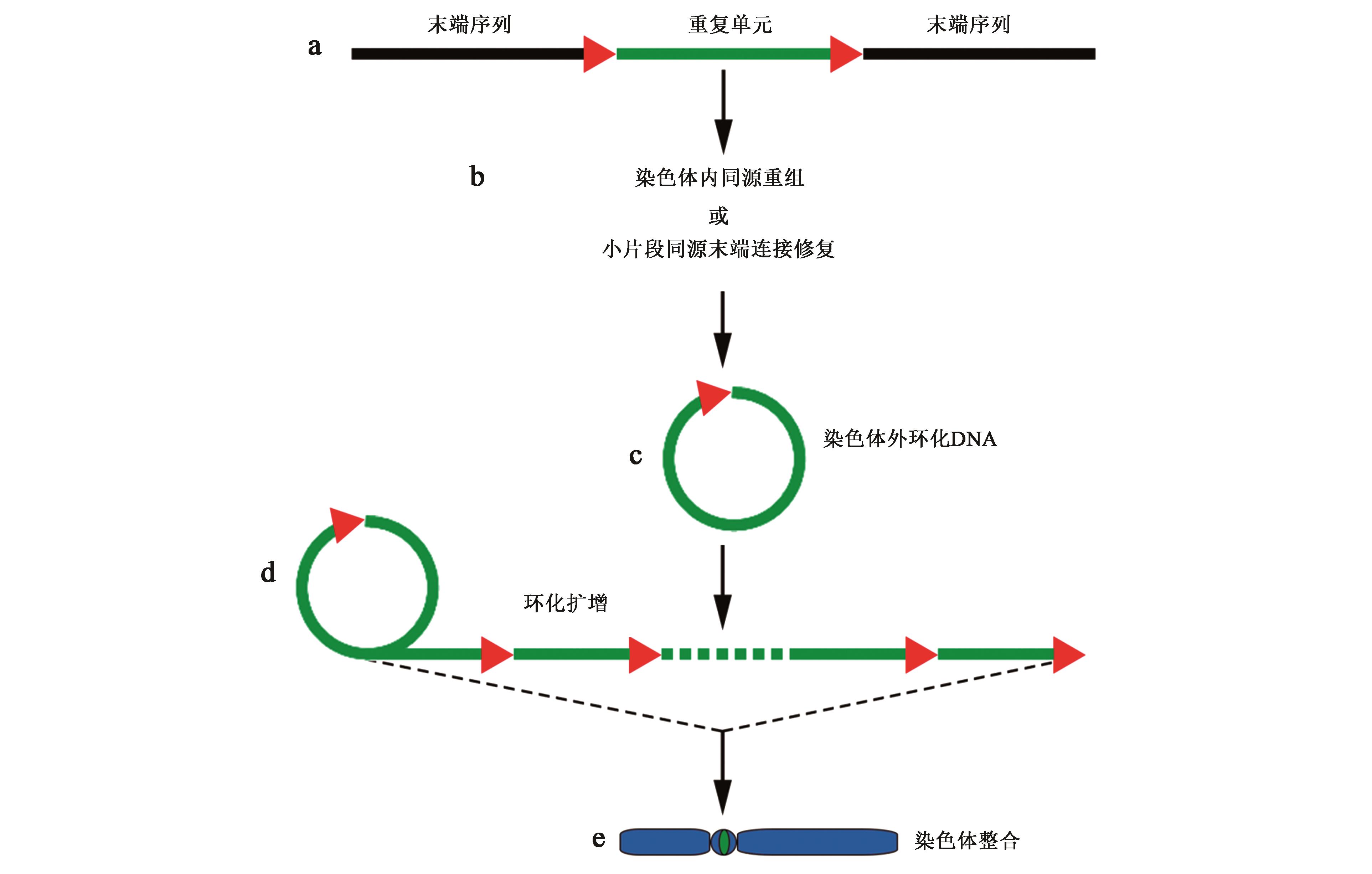
Current Biotechnology ›› 2022, Vol. 12 ›› Issue (1): 1-9.DOI: 10.19586/j.2095-2341.2021.0109
• Reviews • Next Articles
The Research Progress on Plant Functional Centromere DNAs
- 1.School of Life Science,Nantong University,Jiangsu Nantong 226019,China
2.Academic Affairs Office,Nantong University,Jiangsu Nantong 226019,China
-
Received:2021-06-15Accepted:2021-08-05Online:2022-01-25Published:2022-01-26 -
Contact:Kai WANG
植物功能着丝粒DNA研究进展
- 1.南通大学生命科学学院,江苏 南通 226019
2.南通大学教务处,江苏 南通 226019
-
通讯作者:王凯 -
作者简介:张会 E-mail:hzhangfly107@126.com; -
基金资助:国家自然科学基金项目(32070544)
CLC Number:
Cite this article
Hui ZHANG, Kai WANG. The Research Progress on Plant Functional Centromere DNAs[J]. Current Biotechnology, 2022, 12(1): 1-9.
张会, 王凯. 植物功能着丝粒DNA研究进展[J]. 生物技术进展, 2022, 12(1): 1-9.
share this article
Table 1 Known centromeric tandem repeats in plants[23]
| 属名 | 种名 | 重复序列 | 序列大小/bp |
|---|---|---|---|
| 拟南芥属(Arabidopsis) | 大沙叶芥(A. arenosa) | pAa214 | 166~179 |
| 叶芽鼠耳芥(A. gemmifera) | pAge1,pAge2 | 180 | |
| 无苞芥(A. pumila) | pAgKB2,pAgKB1 | 180 | |
| 鼠耳芥(A. thaliana) | pAL1 | 180 | |
| 落花生属(Arachis) | A. batizocoi | AbaTR⁃2 | 297 |
| 落花生(A. hypogaea) | Cot⁃1⁃5,Cot⁃1⁃59,Cot⁃1⁃119 | 115 | |
| A. helodes | AheTR⁃2 | 295 | |
| A. trinitensis | AtrTR⁃2 | 300 | |
| 甜菜属(Beta) | 白花甜菜(B. corolliflora) | pHC8 | 162 |
| 平匍甜菜(B. procumbens) | pTS5,pTS4.1 | 158~312 | |
| 栽培甜菜(B. vulgaris) | pBv1 | 326~327 | |
| 芸薹属(Brassica) | 紫菜薹(B. campestris) | pBcKB4,pBT11 | 175 |
| 结球甘蓝(B. oleracea) | pBoKB1 | 171 | |
| 稻属(Oryza) | 短花药野生稻(O. brachyantha) | CentO⁃F | 154 |
| 根茎野生稻(O. rhizomatis) | CentO⁃C1,CentO⁃C2 | 126,366 | |
| 栽培稻(O. sativa) | CentO | 155 | |
| 茄属(Solanum) | S. bulbocastanum | pSbTC1 | 7 |
| 马铃薯(S. tuberosum) | St18,St24,St49,St57,St3⁃58,St3⁃238,St3⁃294 | 979~5 390 | |
| 紫云英(Astragalus sinicus) | CentAs | 20 | |
| 二穗短柄草(Brachypodium distachyon) | CentBd | 156 | |
| 大豆[Glycine max (soybean)] | CentGm⁃1,CentGm⁃2,CentGm⁃4 | 91~411 | |
| 光叶百脉根(Lotus japonicus) | Ljcen1 | 440 | |
| 紫苜蓿(Medicago sativa) | MsCR | 15~168 | |
| 御谷(Pennisetum glaucum) | pPgKB19 | 137 | |
| 甘蔗热带种(Saccharum officinarum) | SCEN | 140 | |
| 高粱[Sorghum bicolor (sorghum)] | pSau3A10 | 137 | |
| 小麦[Triticum aestivum (wheat)] | Tail family | 570 | |
| 三叶草(Trifolium repens) | TrR350 | 350 | |
| 豇豆(Vigna unguiculata) | pVuKB1 | 488 | |
| 玉米(Zea mays) | CentC,Cent4 | 156,740 | |
Table 1 Known centromeric tandem repeats in plants[23]
| 属名 | 种名 | 重复序列 | 序列大小/bp |
|---|---|---|---|
| 拟南芥属(Arabidopsis) | 大沙叶芥(A. arenosa) | pAa214 | 166~179 |
| 叶芽鼠耳芥(A. gemmifera) | pAge1,pAge2 | 180 | |
| 无苞芥(A. pumila) | pAgKB2,pAgKB1 | 180 | |
| 鼠耳芥(A. thaliana) | pAL1 | 180 | |
| 落花生属(Arachis) | A. batizocoi | AbaTR⁃2 | 297 |
| 落花生(A. hypogaea) | Cot⁃1⁃5,Cot⁃1⁃59,Cot⁃1⁃119 | 115 | |
| A. helodes | AheTR⁃2 | 295 | |
| A. trinitensis | AtrTR⁃2 | 300 | |
| 甜菜属(Beta) | 白花甜菜(B. corolliflora) | pHC8 | 162 |
| 平匍甜菜(B. procumbens) | pTS5,pTS4.1 | 158~312 | |
| 栽培甜菜(B. vulgaris) | pBv1 | 326~327 | |
| 芸薹属(Brassica) | 紫菜薹(B. campestris) | pBcKB4,pBT11 | 175 |
| 结球甘蓝(B. oleracea) | pBoKB1 | 171 | |
| 稻属(Oryza) | 短花药野生稻(O. brachyantha) | CentO⁃F | 154 |
| 根茎野生稻(O. rhizomatis) | CentO⁃C1,CentO⁃C2 | 126,366 | |
| 栽培稻(O. sativa) | CentO | 155 | |
| 茄属(Solanum) | S. bulbocastanum | pSbTC1 | 7 |
| 马铃薯(S. tuberosum) | St18,St24,St49,St57,St3⁃58,St3⁃238,St3⁃294 | 979~5 390 | |
| 紫云英(Astragalus sinicus) | CentAs | 20 | |
| 二穗短柄草(Brachypodium distachyon) | CentBd | 156 | |
| 大豆[Glycine max (soybean)] | CentGm⁃1,CentGm⁃2,CentGm⁃4 | 91~411 | |
| 光叶百脉根(Lotus japonicus) | Ljcen1 | 440 | |
| 紫苜蓿(Medicago sativa) | MsCR | 15~168 | |
| 御谷(Pennisetum glaucum) | pPgKB19 | 137 | |
| 甘蔗热带种(Saccharum officinarum) | SCEN | 140 | |
| 高粱[Sorghum bicolor (sorghum)] | pSau3A10 | 137 | |
| 小麦[Triticum aestivum (wheat)] | Tail family | 570 | |
| 三叶草(Trifolium repens) | TrR350 | 350 | |
| 豇豆(Vigna unguiculata) | pVuKB1 | 488 | |
| 玉米(Zea mays) | CentC,Cent4 | 156,740 | |
Table 2 The centromeric retrotransposons in plants[23]
| 种名 | 重复序列 | 序列大小/bp |
|---|---|---|
| 拟南芥(Arabidopsis thaliana) | Athila | 10 500 |
| 落花生(Arachis hypogaea) | Cot⁃1⁃Clone117(partial) | 117 |
| 平匍甜菜(Beta procumbens) | pBp10 | 417 |
| 甜菜(Beta vulgaris) | pBv26 | 417 |
| 二穗短柄草(Brachypodium distachyon) | CRBd1 | — |
| 大豆[Glycine max (soybean)] | GmCR | 421~852 |
| 雷蒙德棉(Gossypium raimondii) | Gr087,Gr201,Gr285,Gr291,Gr298,Gr304,Gr327,Gr334,Gr344,Gr359,Gr369,Gr372,Gr386,Gr407,Gr442,Gr452,Gr471,Gr552,Gr616 | 309~5 909 |
| 大麦(Hordeum vulgare) | cereba | 7 176 |
| 番茄(Lycopersicum esculentum) | TGRIV | 7 000 |
| 水稻(Oryza sativa) | CRR | 7 400~7 800 |
| 黑麦(Secale cereale) | Bilby | 3 400 |
| 甘蔗热带种(Saccharum officinarum) | CRS | 3 600 |
| 高粱(Sorghum bicolor) | pSau3A9 | 745 |
| 野生一粒小麦(Triticum boeoticum) | CRWs | 7 762~7 865 |
| 玉米(Zea mays) | CRM | 7 572 |
| Zingeria biebersteiniana | Zb47A | 263 |
Table 2 The centromeric retrotransposons in plants[23]
| 种名 | 重复序列 | 序列大小/bp |
|---|---|---|
| 拟南芥(Arabidopsis thaliana) | Athila | 10 500 |
| 落花生(Arachis hypogaea) | Cot⁃1⁃Clone117(partial) | 117 |
| 平匍甜菜(Beta procumbens) | pBp10 | 417 |
| 甜菜(Beta vulgaris) | pBv26 | 417 |
| 二穗短柄草(Brachypodium distachyon) | CRBd1 | — |
| 大豆[Glycine max (soybean)] | GmCR | 421~852 |
| 雷蒙德棉(Gossypium raimondii) | Gr087,Gr201,Gr285,Gr291,Gr298,Gr304,Gr327,Gr334,Gr344,Gr359,Gr369,Gr372,Gr386,Gr407,Gr442,Gr452,Gr471,Gr552,Gr616 | 309~5 909 |
| 大麦(Hordeum vulgare) | cereba | 7 176 |
| 番茄(Lycopersicum esculentum) | TGRIV | 7 000 |
| 水稻(Oryza sativa) | CRR | 7 400~7 800 |
| 黑麦(Secale cereale) | Bilby | 3 400 |
| 甘蔗热带种(Saccharum officinarum) | CRS | 3 600 |
| 高粱(Sorghum bicolor) | pSau3A9 | 745 |
| 野生一粒小麦(Triticum boeoticum) | CRWs | 7 762~7 865 |
| 玉米(Zea mays) | CRM | 7 572 |
| Zingeria biebersteiniana | Zb47A | 263 |
| 1 | BLOOM K S. Centromeric heterochromatin: the primordial segregation machine [J]. Annu. Rev. Genet., 2014, 48(1): 457-484. |
| 2 | AHMAD S F, SINGCHAT W, JEHANGIR M, et al.. Dark matter of primate genomes: satellite DNA repeats and their evolutionary dynamics [J/OL]. Cells, 2020, 9: 2714[2020-12-24]. . |
| 3 | BIGGINS S. The composition, functions, and regulation of the budding yeast kinetochore [J]. Genetics, 2013, 194(4): 817-846. |
| 4 | VARMA D, WAN X, CHEERAMBATHUR D, et al.. Spindle assembly checkpoint proteins are positioned close to core microtubule attachment sites at kinetochores [J]. J. Cell Biol., 2013, 202(5): 735-746. |
| 5 | WESTHORPE F G, STRAIGHT A F. Functions of the centromere and kinetochore in chromosome segregation [J]. Curr. Opin. Cell Biol., 2013, 25(3): 334-340. |
| 6 | PRESTING G. Centromeric retrotransposons and centromere function [J]. Curr. Opin. Genet. Dev., 2018, 49: 79-84. |
| 7 | MALIK H S, HENIKOFF S. Major evolutionary transitions in centromere complexity [J]. Cell, 2009, 138(6): 1067-1082. |
| 8 | LERMONTOVA I, KOROLEVA O, RUTTEN T, et al.. Knockdown of CENH3 in Arabidopsis reduces mitotic divisions and causes sterility by disturbed meiotic chromosome segregation [J]. Plant J., 2011, 68(1): 40-50. |
| 9 | TALBERT P B, MASUELLI R, TYAGI A P, et al.. Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant [J]. Plant Cell, 2002, 14(5): 1053-1066. |
| 10 | NAGAKI K, CHENG Z, OUYANG S, et al.. Sequencing of a rice centromere uncovers active genes [J]. Nat. Genet., 2004, 36(2): 138-145. |
| 11 | TEK A L, KASHIHARA K, MURATA M, et al.. Identification of the centromere-specific histone H3 variant in Lotus japonicus [J]. Gene, 2014, 538(1): 8-11. |
| 12 | KIXMOELLER K, ALLU P K, BLACK B E. The centromere comes into focus: from CENP-A nucleosomes to kinetochore connections with the spindle [J/OL] . Open Biol., 2020, 10(6): 200051[2020-06-10]. . |
| 13 | MADDOX P S, PORTIER N, DESAI A, et al.. Molecular analysis of mitotic chromosome condensation using a quantitative time-resolved fluorescence microscopy assay [J]. Proc. Natl. Acad. Sci. USA, 2006, 103(41): 15097-15102. |
| 14 | VENKEI Z, PRZEWLOKA M R, LADAK Y, et al.. Spatiotemporal dynamics of Spc105 regulates the assembly of the Drosophila kinetochore [J/OL] . Open Biol., 2012, 2(2): 110032[2012-02-01]. . |
| 15 | OLIVEIRA L C, TORRES G A. Plant centromeres: genetics, epigenetics and evolution [J]. Mol. Biol. Rep., 2018, 45:1491-1497. |
| 16 | MELTERS D P, BRADNAM K R, YOUNG H A, et al.. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution [J/OL]. Genome Biol., 2013, 14(1): R10[2013-01-30]. . |
| 17 | COPENHAVER G P, NICKEL K, KUROMORI T, et al.. Genetic definition and sequence analysis of Arabidopsis centromeres [J]. Science, 1999, 286(5449): 2468-2474. |
| 18 | GAO D, GILL N, KIM H R, et al.. A lineage-specific centromere retrotransposon in Oryza brachyantha [J]. Plant J., 2009, 60(5): 820-831. |
| 19 | ZHANG H, KOBLIZKOVA A, WANG K, et al.. Boom-bust turnovers of megabase-sized centromeric dna in Solanum species: rapid evolution of dna sequences associated with centromeres [J]. Plant Cell, 2014, 26(4): 1436-1447. |
| 20 | WU Y, KIKUCHI S, YAN H, et al.. Euchromatic subdomains in rice centromeres are associated with genes and transcription [J]. Plant Cell, 2011, 23(11): 4054-4064. |
| 21 | GONG Z, WU Y, KOBLIZKOVA A, et al.. Repeatless and repeat-based centromeres in potato: implications for centromere evolution [J]. Plant Cell, 2012, 24(9): 3559-3574. |
| 22 | HUANG Y, DING W, ZHANG M, et al.. The formation and evolution of centromeric satellite repeats in Saccharum species [J]. Plant J., 2021, 106(3): 616-629. |
| 23 | 欧阳凯.芸薹属植物功能着丝粒序列组成及进化分析[D]. 福州:福建农林大学,2020. |
| 24 | LI Y, ZUO S, ZHANG Z, et al.. Centromeric DNA characterization in the model grass Brachypodium distachyon provides insights on the evolution of the genus [J]. Plant J., 2018, 93(6): 1088-1101. |
| 25 | HIATT E N, KENTNER E K, Dawe R K. Independently regulated neocentromere activity of two classes of tandem repeat arrays [J]. Plant Cell, 2002, 14(2): 407-420. |
| 26 | ZHANG W, ZUO S, LI Z, et al.. Isolation and characterization of centromeric repetitive DNA sequences in Saccharum spontaneum [J/OL]. Sci. Rep., 2017, 7: 41659[2017-01-31]. . |
| 27 | CHENG Z, DONG F, LANGDON T, et al.. Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon [J]. Plant Cell, 2002, 14(8): 1691-1704. |
| 28 | LEE H R, ZHANG W, LANGDON T, et al.. Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species [J]. Proc. Natl. Acad. Sci. USA, 2005, 102(33): 11793-11798. |
| 29 | YANG X, ZHAO H, ZHANG T, et al.. Amplification and adaptation of centromeric repeats in polyploid switchgrass species [J]. New Phytol., 2018, 218(4): 1645-1657. |
| 30 | LAMB J C, BIRCHLER J A. The role of DNA sequence in centromere formation [J/OL]. Genome Biol., 2003, 4(5): 214[2003-04-29]. . |
| 31 | ZHONG C X, MARSHALL J B, TOPP C, et al.. Centromeric retroelements and satellites interact with maize kinetochore protein CENH3 [J]. Plant Cell, 2002, 14(11): 2825-2836. |
| 32 | JIANG J, BIRCHLER J A, PARROTT W A, et al.. A molecular view of plant centromeres[J]. Trends Plant Sci., 2003, 8(12): 570-575. |
| 33 | FENG Q, ZHANG Y J, HAO P, et al.. Sequence and analysis of rice chromosome 4 [J]. Nature, 2002, 420(6913): 316-320. |
| 34 | ANANIEV E V, PHILLIPS R L, RINES H W. Chromosome-specific molecular organization of maize (Zea mays L.) centromeric regions [J]. Proc. Natl. Acad. Sci. USA, 1998, 95(22): 13073-13078. |
| 35 | AMBORELLA G P. The Amborella genome and the evolution of flowering plants [J]. Science, 2013, 342(6165): 1241089[2013-12-20]. . |
| 36 | JIANG J, BIRCHLER J A. Plant Centromere Biology [M]. USA: Wiley-Blackwell, 2013. |
| 37 | HAN J, MASONBRINK R E, SHAN W, et al.. Rapid proliferation and nucleolar organizer targeting centromeric retrotransposons in cotton [J]. Plant J., 2016, 88(6): 992-1005. |
| 38 | ZHU Z, GUI S, JIN J, et al.. The NnCenH3 protein and centromeric DNA sequence profiles of Nelumbo nucifera Gaertn. (Sacred lotus) reveal the DNA structures and dynamics of centromeres in basal eudicots[J]. Plant J., 2016, 87(6): 568-582. |
| 39 | LI B, CHOULET F, HENG Y, et al.. Wheat centromeric retrotransposons: the new ones take a major role in centromeric structure [J]. Plant J., 2013, 73(6): 952-965. |
| 40 | YAN H, JIN W, NAGAKI K, et al.. Transcription and histone modifications in the recombination-free region spanning a rice centromere [J]. Plant Cell, 2005, 17(12): 3227-3238. |
| 41 | SUN X, WAHLSTROM J, KARPEN G. Molecular structure of a functional Drosophila centromere[J]. Cell, 1997, 91(7): 1007-1019. |
| 42 | WU J, YAMAGATA H, HAYASHI-TSUGANE M, et al.. Composition and structure of the centromeric region of rice chromosome 8 [J]. Plant Cell, 2004, 16(4): 967-976. |
| 43 | WANG K, WU Y, ZHANG W, et al.. Maize centromeres expand and adopt a uniform size in the genetic background of oat [J]. Genome Res., 2014, 24(1): 107-116. |
| 44 | IOVENE M, YU Q, MING R, et al.. Evidence for emergence of sex-determining gene(s) in a centromeric region in Vasconcellea parviflora [J]. Genetics, 2015, 199(2): 413-421. |
| 45 | YOU Q, ZHAI K, YANG D, et al.. An E3 ubiquitin ligase-BAG protein module controls plant innate immunity and broad-spectrum disease resistance [J]. Cell Host Microbe, 2016, 20(6): 758-769. |
| 46 | XING J, NING Q, TANG D, et al.. Progress on the role of extrachromosomal DNA in tumor pathogenesis and evolution [J]. Clin. Genet., 2021, 99(4): 503-512. |
| 47 | TORRES G A, GONG Z, IOVENE M, et al.. Organization and evolution of subtelomeric satellite repeats in the potato genome [J]. G3 (Bethesda), 2011, 1(2): 85-92. |
| 48 | HE L, LIU J, TORRES G A, et al.. Interstitial telomeric repeats are enriched in the centromeres of chromosomes in Solanum species [J]. Chrom. Res., 2013, 21(1): 5-13. |
| 49 | ALFREDO V, MÉNDEZ-LAGO M, ABAD J P, et al.. The birth of the centromere [J]. Cell Cycle (Georgetown, Tex.), 2007, 6(23): 2872-2876. |
| 50 | ZHANG T, TALBERT P B, ZHANG W, et al.. The CentO satellite confers translational and rotational phasing on cenH3 nucleosomes in rice centromeres [J]. Proc. Natl. Acad. Sci. USA, 2013, 110(50): 4875-4883. |
| 51 | HAN M, YANG Y, ZHANG M, et al.. Considerations regarding centromere assembly in plant whole-genome sequencing [J]. Methods, 2021, 187: 54-56. |
| 52 | HU Y, CHEN J, FANG L, et al.. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton [J]. Nat. Genet., 2019, 51(4): 739-748. |
| 53 | NOVAK P, NEUMANN P, MACAS J. Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data [J/OL]. BMC Bioinform., 2010, 11: 378[2010-07-15]. . |
| [1] | Zhenhua KE. Research on Detection Methods of Pseudomonas aeruginosa in Cold Chain Food and Construction of Evolutionary Tree [J]. Current Biotechnology, 2023, 13(6): 940-944. |
| [2] | Weiping ZHANG, Bingying QIU, Dongsheng ZHANG. Adaptive Evolution Analysis of Mitochondrial Genomes in Anseriform Birds with Various Feeding Habits [J]. Current Biotechnology, 2023, 13(5): 748-754. |
| [3] | ZHENG Qi-lian1,2, GUO Ya-jie2, LIN Qian-nan2, CHEN Xiao-gang3, LI Zong-di3, DAI Xiao-yi3. Progress of the Natural Evolution and Action Mechanism of Cry Toxin [J]. Curr. Biotech., 2016, 6(1): 14-18. |
| [4] | GUO Ya-jie1,2, LI Ping1, WU Zhong-ling1, ZHENG Ru-ping1, ZHANG Jun1,2, HU Xia1*. Progress on Molecular Modification Methods of Insecticidal Activity of Cry Toxin [J]. Curr. Biotech., 2016, 6(1): 19-24. |
| [5] | HE Zhen1, YUN Xiao-dong1, WU Kai1, JI Hu-tai2, CHANG Jian-zhong1, QIAO Lin-yi1*, ZHENG Jun2*. Progress on Germplasm Resources Genetic Diversity of Soybean [J]. Curr. Biotech., 2015, 5(2): 103-108. |
| [6] | LUAN Guo-dong, CAI Zhen, LI Yin*. Development and Application of Hypermutable Cells in Evolutionary Engineering [J]. Curr. Biotech., 2014, 4(4): 229-235. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||