Current Biotechnology ›› 2021, Vol. 11 ›› Issue (4): 518-525.DOI: 10.19586/j.2095-2341.2020.0144
• Human Health and the Environment • Previous Articles Next Articles
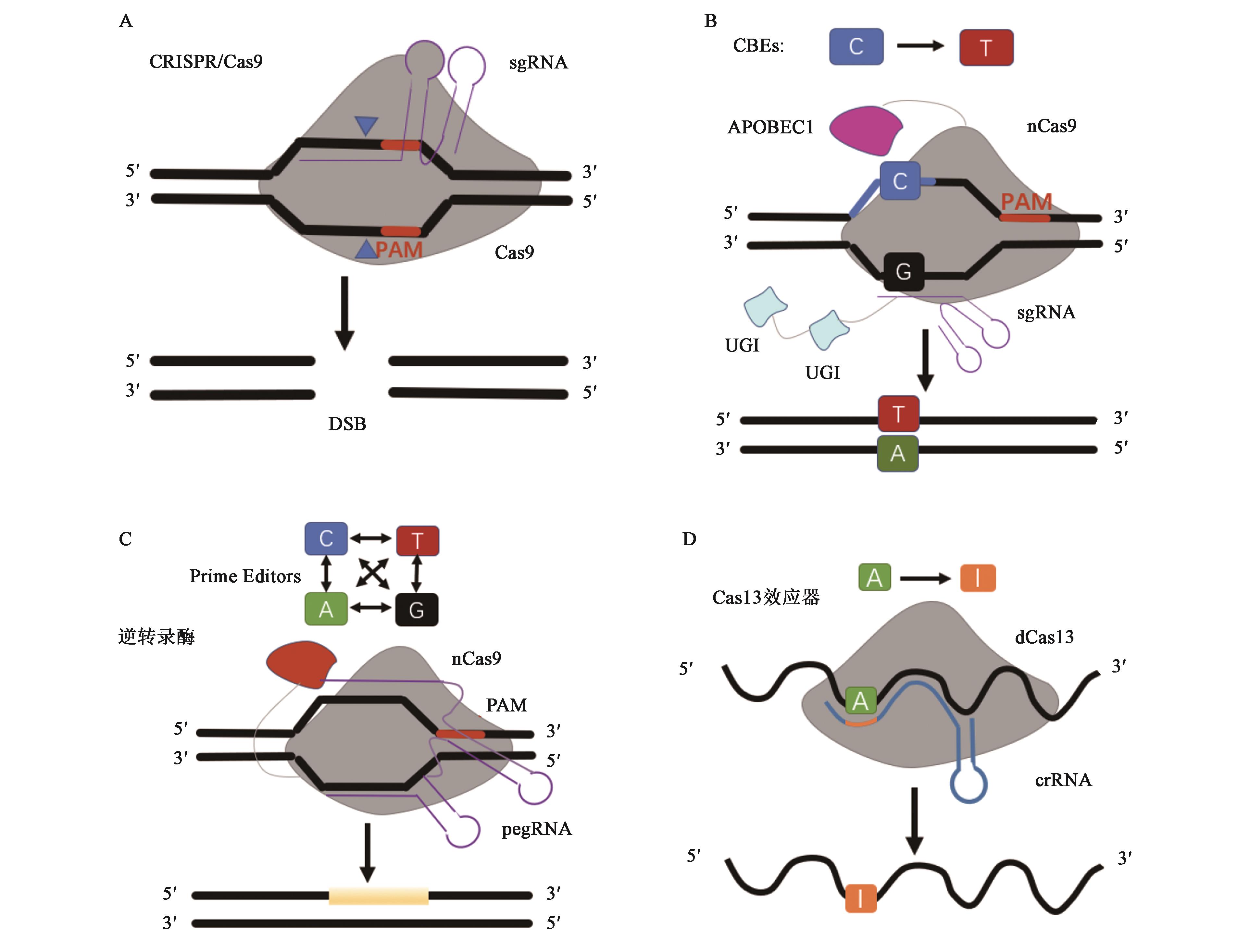
Application Progress of CRISPR/Cas and its Derivative Editing Technology in Gene Therapy
Chunju QUAN( ), Zhongliang ZHENG(
), Zhongliang ZHENG( )
)
- College of Life Sciences,Wuhan University,Wuhan 430027,China
-
Received:2020-11-06Accepted:2021-03-04Online:2021-07-25Published:2021-08-02 -
Contact:Zhongliang ZHENG
CRISPR/Cas及其衍生编辑技术在基因治疗中的应用进展
- 武汉大学生命科学学院,武汉 430027
-
通讯作者:郑忠亮 -
作者简介:权春菊 E-mail:1096150987@qq.com; -
基金资助:国家自然科学基金项目(81372441)
CLC Number:
Cite this article
Chunju QUAN, Zhongliang ZHENG. Application Progress of CRISPR/Cas and its Derivative Editing Technology in Gene Therapy[J]. Current Biotechnology, 2021, 11(4): 518-525.
权春菊, 郑忠亮. CRISPR/Cas及其衍生编辑技术在基因治疗中的应用进展[J]. 生物技术进展, 2021, 11(4): 518-525.
share this article
| 1 | DOUDNA J A. The promise and challenge of therapeutic genome editing[J]. Nature, 2020, 578(7794): 229-236. |
| 2 | TAMAS L, YANG D, WALDMAN A S. Repair of a specific double-strand break generated within a mammalian chromosome by yeast endonuclease I-SceI [J]. Nucl. Acids Res., 1994, 22(25): 5649-5657. |
| 3 | WYMAN C, KANAAR R. DNA double-strand break repair: all's well that ends well [J]. Annu. Rev. Genet., 2006, 40(1):363-383. |
| 4 | URNOV F D, REBAR E J, HOLMES M C, et al.. Genome editing with engineered zinc finger nucleases[J]. Nat. Rev. Genet., 2010, 11(9): 636-646. |
| 5 | JOUNG J K, SANDER J D. TALENs: a widely applicable technology for targeted genome editing [J]. Nat. Rev. Mol. Cell Biol., 2012, 14(1): 49-55. |
| 6 | CHO S W, KIM S, KIM J M, et al.. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease [J]. Nat. Biotechnol., 2013, 31(3): 230-232. |
| 7 | CONG L, RAN F A, COX D M, et al.. Multiplex genome engineering using CRISPR/Cas systems [J]. Science, 2013, 339(6121): 819-823. |
| 8 | MALI P, YANG L H, ESVELT K M, et al.. RNA-guided human genome engineering via Cas9 [J]. Science, 2013, 339(6121): 823-826. |
| 9 | MAKAROVA K S, WOLF Y I, ALKHNBASHI O S, et al.. An updated evolutionary classification of CRISPR-Cas systems [J]. Nat. Rev. Microbiol., 2015, 13(11): 722-736. |
| 10 | MOHANRAJU P, MAKAROVA K S, ZETSCHE B, et al.. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems [J/OL]. Science, 2016, 353(6299): aad5147[2021-06-04]. . DOI: 10.1126/science.aad5147 . |
| 11 | ZEBALLOS C M A, GAJ T. Next-generation CRISPR technologies and their applications in gene and cell therapy [J/OL]. Trends Biotechnol., 2020, 1: S0167-7799(20)30287-0[2021-06-04]. . |
| 12 | WANG F, WANG L, ZOU X, et al.. Advances in CRISPR-Cas systems for RNA targeting, tracking and editing[J]. Biotechnol. Adv., 2019, 37(5): 708-729. |
| 13 | 任云晓, 肖茹丹, 娄晓敏, 等. 基因编辑技术及其在基因治疗中的应用[J]. 遗传, 2019, 41(1): 18-27. |
| 14 | JINEK M, CHYLINSKI K, FONFARA I, et al.. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity [J]. Science, 2012, 337(6096): 816-821. |
| 15 | KOMOR A C, KIM Y B, PACKER M S, et al.. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage [J]. Nature, 2016, 533(7603): 420-424. |
| 16 | NISHIDA K, ARAZOE T, YACHIE N, et al.. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems [J/OL]. Science, 2016, 353(6305): aaf8729[2021-06-04]. . DOI: 10.1126/science.aaf8729 . |
| 17 | GAUDELLI N M, KOMOR A C, REES H A, et al.. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage [J]. Nature, 2017, 551(7681): 464-471. |
| 18 | ANZALONE A V, RANDOLPH P B, DAVIS J R, et al.. Search-and-replace genome editing without double-strand breaks or donor DNA [J]. Nature, 2019, 576(7785): 149-157. |
| 19 | YEH W H, CHIANG H, REES H A, et al.. In vivo base editing of post-mitotic sensory cells [J/OL]. Nat. Commun., 2018, 9(1): 2184[2021-06-04]. . |
| 20 | VILLIGER L, GRISCH-CHAN H M, LINDSAY H, et al.. Treatment of a metabolic liver disease by in vivo genome base editing in adult mice [J]. Nat. Med., 2018, 24(10): 1519-1525. |
| 21 | LEVY J M, YEH W H, PENDSE N, et al.. Cytosine and adenine base editing of the brain, liver, retina, heart and skeletal muscle of mice via adeno-associated viruses [J]. Nat. Biomed. Eng., 2020, 4(1): 97-110. |
| 22 | LIM C K W, GAPINSKE M, BROOKS A K, et al.. Treatment of a mouse model of ALS by in vivo base editing [J]. Mol. Ther., 2020, 28(4): 1177-1189. |
| 23 | WINTER J, LUU A, GAPINSKE M, et al.. Targeted exon skipping with AAV-mediated split adenine base editors [J/OL]. Cell Discov., 2019, 5:41[2021-06-04]. . |
| 24 | ABUDAYYEH O O, GOOTENBERG J S, ESSLETZBICHLER P, et al.. RNA targeting with CRISPR-Cas13 [J]. Nature, 2017, 550(7675): 280-284. |
| 25 | ABUDAYYEH O O, GOOTENBERG J S, KONERMANN S, et al.. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector [J/OL]. Science, 2016, 353(6299): aaf5573[2021-06-04]. . DOI: 10.1126/science.aaf5573 . |
| 26 | EAST-SELETSKY A, O'CONNELL M R, KNIGHT S C, et al.. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection [J]. Nature, 2016, 538(7624): 270-273. |
| 27 | MEESKE A J, NAKANDAKARI-HIGA S, MARRAFFINI L A. Cas13-induced cellular dormancy prevents the rise of CRISPR-resistant bacteriophage [J]. Nature, 2019, 570(7760): 241-245. |
| 28 | SMARGON A A, COX D B T, PYZOCHA N K, et al.. Cas13b is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28 [J]. Mol. Cell, 2017, 65(4):618-630. |
| 29 | PARK S H, LEE C M, DEVER D P, et al.. Highly efficient editing of the beta-globin gene in patient-derived hematopoietic stem and progenitor cells to treat sickle cell disease [J]. Nucleic Acids Res., 2019, 47(15): 7955-7972. |
| 30 | KALKAN B M, KALA E Y, YUCE M, et al.. Development of gene editing strategies for human beta-globin (HBB) gene mutations [J/OL]. Gene, 2020, 734:144398[2021-06-04]. . |
| 31 | DEVER D P, BAK R O, REINISCH A, et al.. CRISPR/Cas9 beta-globin gene targeting in human haematopoietic stem cells [J]. Nature, 2016, 539(7629): 384-389. |
| 32 | KOSICKI M, TOMBERG K, BRADLEY A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements [J]. Nat. Biotechnol., 2018, 36(8): 765-771. |
| 33 | RYU S M, KOO T, KIM K, et al.. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy [J]. Nat. Biotechnol., 2018, 36(6): 536-539. |
| 34 | REN J, ZHAO Y. Advancing chimeric antigen receptor T cell therapy with CRISPR/Cas9 [J]. Protein Cell, 2017, 8(9): 634-643. |
| 35 | ZHAO J, LIN Q, SONG Y, et al.. Universal CARs, cellsuniversal T, and universal CAR T cells [J/OL]. J. Hematol. Oncol., 2018, 11(1): 132[2021-06-04]. . |
| 36 | LU Y, XUE J, DENG T, et al.. Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer [J]. Nat. Med., 2020, 26(5): 732-740. |
| 37 | LIU D, ZHAO X, TANG A, et al.. CRISPR screen in mechanism and target discovery for cancer immunotherapy [J/OL]. Biochim. Biophys. Acta Rev. Cancer, 2020, 1874(1): 188378[2021-06-04]. . |
| 38 | CYRANOSKI D. Chinese scientists to pioneer first human CRISPR trial [J]. Nature, 2016, 535(7613): 476-477. |
| 39 | CYRANOSKI D. CRISPR gene-editing tested in a person for the first time [J]. Nature, 2016, 539(7630): 479. |
| 40 | STADTMAUER E A, FRAIETTA J A, DAVIS M M, et al.. CRISPR-engineered T cells in patients with refractory cancer [J/OL]. Science, 2020, 367(6481):eaba7365[2021-06-04]. . DOI: 10.1126/science.aba7365 . |
| 41 | ZAFRA M P, SCHATOFF E M, KATTI A, et al.. Optimized base editors enable efficient editing in cells, organoids and mice [J]. Nat. Biotechnol., 2018, 36(9): 888-893. |
| 42 | ZHAO X, LIU L, LANG J, et al.. A CRISPR-Cas13a system for efficient and specific therapeutic targeting of mutant KRAS for pancreatic cancer treatment [J]. Cancer Lett., 2018, 431:171-181. |
| 43 | WANG E, ZHOU H, NADORP B, et al.. Surface antigen-guided CRISPR screens identify regulators of myeloid leukemia differentiation [J]. Cell Stem Cell, 2021, 28(4):718-731. |
| 44 | ZHU W, XIE K, XU Y, et al.. CRISPR/Cas9 produces anti-hepatitis B virus effect in hepatoma cells and transgenic mouse [J]. Virus Res., 2016, 217:125-132. |
| 45 | ROSSIDIS A C, STRATIGIS J D, CHADWICK A C, et al.. In utero CRISPR-mediated therapeutic editing of metabolic genes [J]. Nat. Med., 2018, 24(10): 1513-1518. |
| 46 | ZHANG X, ZHAO W, NGUYEN G N, et al.. Functionalized lipid-like nanoparticles for in vivo mRNA delivery and base editing [J/OL]. Sci. Adv., 2020, 6(34):eabc2315[2021-06-04]. . DOI: 10.1126/sciadv.abc2315 . |
| 47 | CHADWICK A C, WANG X, MUSUNURU K. In vivo base editing of PCSK9 (proprotein convertase subtilisin/kexin type 9) as a therapeutic alternative to genome editing [J]. Arterioscler. Thromb. Vasc. Biol., 2017, 37(9): 1741-1747. |
| 48 | ZHOU H, SU J, HU X, et al.. Glia-to-neuron conversion by CRISPR-CasRx alleviates symptoms of neurological disease in mice [J]. Cell, 2020, 181(3): 590-603. |
| 49 | KOBLAN L W, DOMAN J L, WILSON C, et al.. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction [J]. Nat. Biotechnol., 2018, 36(9): 843-846. |
| 50 | LIANG P, XIE X, ZHI S, et al.. Genome-wide profiling of adenine base editor specificity by EndoV-seq [J/OL]. Nat. Commun., 2019, 10(1): 67[2021-06-04]. . |
| 51 | REES H A, KOMOR A C, YEH W H, et al.. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery [J/OL]. Nat. Commun., 2017, 8(1):15790[2021-06-04]. . |
| 52 | GRüNEWALD J, ZHOU R, GARCIA S P, et al.. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors [J]. Nature, 2019, 569(7756): 433-437. |
| 53 | JIANG W, FENG S, HUANG S, et al.. BE-PLUS: a new base editing tool with broadened editing window and enhanced fidelity [J]. Cell Res., 2018, 28(8): 855-861. |
| 54 | GAUDELLI N M, LAM D K, REES H A, et al.. Directed evolution of adenine base editors with increased activity and therapeutic application [J]. Nat. Biotechnol., 2020, 38(7): 892-900. |
| 55 | ZHANG X, CHEN L, ZHU B, et al.. Increasing the efficiency and targeting range of cytidine base editors through fusion of a single-stranded DNA-binding protein domain [J]. Nat. Cell Biol., 2020, 22(6): 740-750. |
| 56 | JVAN HAASTEREN, LI J, SCHEIDELER O J, et al.. The delivery challenge: fulfilling the promise of therapeutic genome editing [J]. Nat. Biotechnol., 2020, 38(7): 845-855. |
| 57 | MENDELL J R, AL-ZAIDY S, SHELL R, et al.. Single-dose gene-replacement therapy for spinal muscular atrophy [J]. N. Engl. J. Med., 2017, 377(18): 1713-1722. |
| 58 | Goldschmidt D, Scutti S. FDA approves gene therapy for a type of blindness[EB/OL]. (2017-12-21) [2021-06-29]. . |
| 59 | ALANIS-LOBATO G, ZOHREN J, MCCARTHY A, et al.. Frequent loss-of-heterozygosity in CRISPR-Cas9-edited early human embryos [J/OL]. Proc. Natl. Acad. Sci. USA, 2021, 118(22): e2004832117 [2021-06-04]. . |
| 60 | ZUCCARO M V, XU J, MITCHELL C, et al.. Reading frame restoration at the EYS locus, and allele-specific chromosome removal after Cas9 cleavage in human embryos [J]. Cell, 2020, 183(6):1650-1664. |
| 61 | CHARLESWORTH C T, DESHPANDE P S, DEVER D P, et al.. Identification of preexisting adaptive immunity to Cas9 proteins in humans [J]. Nat. Med., 2019, 25(2): 249-254. |
| [1] | Bingyue QIU, Jiateng SHI, Jing JIN. Research Progress of siRNA Gene Therapy in Tendon Injury Repair [J]. Current Biotechnology, 2025, 15(3): 411-417. |
| [2] | Yiyang LI, Zhizheng ZHOU, Shufei WANG, Boya LIU, Yufei LIU, Xiaoyan LI, Hongshu SUI, Dongwei LIU. Application and Prospect of CRISPR/Cas9 Gene Editing Technology in Disease Treatment [J]. Current Biotechnology, 2025, 15(1): 35-42. |
| [3] | Guang HU, Zhi WANG, Wei FU, Yuting SHI, Shanshan CHEN, Liang LUO, Shuang WEI. Establishment of Detection Method Based on TaqMan Real-time Fluorescence Quantitative PCR Technology for OsWx-edited Rice [J]. Current Biotechnology, 2025, 15(1): 86-92. |
| [4] | Jing WANG, Haitao GUAN, Xiaolei ZHANG, Baohuai WANG, Baohai LIU, Hongtao WEN. Detection Dynamic and Development Tendency of Agricultural Gene Editing Products [J]. Current Biotechnology, 2024, 14(5): 712-723. |
| [5] | Hongkai WANG, Yujie WANG, Xiaohan ZHAO, Yuhan JING, Jiayi TANG, Hongshu SUI. Research Progress on Cystic Fibrosis Gene Therapy [J]. Current Biotechnology, 2024, 14(5): 813-819. |
| [6] | Mingyang JIA, Lei WANG, Junfeng CHEN, Jiaqing ZHANG, Xiangzhou YAN, Baosong XING, Jing WANG. Research Progress of CRISPR/Cas9 Gene Editing Technology in Livestock and Poultry Breeding [J]. Current Biotechnology, 2024, 14(4): 529-536. |
| [7] | Jiacong ZHANG, Jigang LU. Establishment of Biallelic Knockout Technique in Nile Tilapia (Oreochromis niloticus) Based on CRISPR/Cas9 System: A Case Study of SLC24A5 Gene [J]. Current Biotechnology, 2024, 14(3): 442-450. |
| [8] | Xiaotian ZHANG, Zhi WANG, Pengyu ZHU, Shuang WEI, Wei FU, Chunmeng HUANG, Zhihong LI, Huiyu WANG, Yue JIAO. A Rapid Detection Method Based on qPCR for CRISPR/Cas9 Edited Crops [J]. Current Biotechnology, 2023, 13(6): 907-912. |
| [9] | Ali WANG, Jiangdong LIU. Research Progress on the CRISPR/Cas System in Zebrafish [J]. Current Biotechnology, 2023, 13(4): 485-491. |
| [10] | Maolan XIONG, Siyan WEI, Juntao LUO, Bingshe HAN, Junfang ZHANG. The Effects of hdac11 Knockout of Zebrafish on Lipid Metabolism [J]. Current Biotechnology, 2023, 13(4): 588-595. |
| [11] | Kun YU, Jiaqi XUE, Jinkuan WANG, Yongtao YU. Research Progress on Application of CRISPR/Cas9 Gene Editing Technique in Filamentous Fungi [J]. Current Biotechnology, 2022, 12(5): 696-704. |
| [12] | Weisong GAO, Jinping DOU, Shuang WEI, Xingjian LIU, Zhifang ZHANG, Yinyu LI. Classification and Research Status of CRISPR/Cas Systems [J]. Current Biotechnology, 2022, 12(4): 532-538. |
| [13] | Yunyan FEI, Jun YANG, Dedao JING, Tianzi LIN, Chuang LI, Huafei QIAN, Shengyuan ZENG, Huaxin HAN, Hongbing GONG. Research and Application Progress of CRISPR/Cas Technology in Herbicide⁃resistant Crops Breeding [J]. Current Biotechnology, 2022, 12(2): 189-197. |
| [14] | Shuxiang LI, Liping AN, Dejuan LIANG, Kaixuan WANG, Jianguo ZHAO. Research Progress on the Treatment of Prion Disease [J]. Current Biotechnology, 2022, 12(2): 229-235. |
| [15] | Qiao CAO, Zhanliang SHI, Guocong ZHANG, Jinfu BAN, Shusong ZHENG, Xiaoyi FU, Shichang ZHANG, Mingqi HE, Ran HAN, Zhenxian GAO. Progress of CRISPR/Cas9 Application in Wheat Breeding [J]. Current Biotechnology, 2021, 11(6): 661-667. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||