Current Biotechnology ›› 2025, Vol. 15 ›› Issue (3): 544-554.DOI: 10.19586/j.2095-2341.2024.0195
• Articles • Previous Articles
Genetic Polymorphism Analysis on Short Tandem Repeats of Y Chromosome in Han Population of Hohhot
Luyao LI1( ), Ning LYU2(
), Ning LYU2( ), Yongjie ZHAI2, Run ZHANG3, Anxin YAN4, Fuquan JIA5(
), Yongjie ZHAI2, Run ZHANG3, Anxin YAN4, Fuquan JIA5( )
)
- 1.Department of Pathology,The First Affiliated Hospital of Xi'an Jiaotong University,Xi'an 710061,China
2.Hohhot Public Security Bureau,Hohhot 010051,China
3.Chifeng Public Security Bureau,Inner Mongolia Chifeng 024000,China
4.Institute of Identification,Ministry of Public Security,Beijing 100038,China
5.The Department of Forensic Medicine,Inner Mongolia Medical University,Hohhot 010059,China.
-
Received:2024-12-18Accepted:2025-02-12Online:2025-05-25Published:2025-07-01 -
Contact:Fuquan JIA
呼和浩特汉族人群Y-STR的遗传多态性分析
李璐瑶1( ), 吕宁2(
), 吕宁2( ), 翟永结2, 张润3, 严安心4, 贾富全5(
), 翟永结2, 张润3, 严安心4, 贾富全5( )
)
- 1.西安交通大学第一附属医院病理科,西安 710061
2.呼和浩特市公安局,呼和浩特 010051
3.赤峰市公安局,内蒙古 赤峰 024000
4.公安部鉴定中心,北京 100038
5.内蒙古医科大学基础医学院法医学教研室,呼和浩特 010059
-
通讯作者:贾富全 -
作者简介:李璐瑶 E-mail: luyaoli@xjtufh.edu.cn
吕宁 E-mail: 13948514862@139.com第一联系人:(李璐瑶、吕宁并列第一作者) -
基金资助:内蒙古医科大学大学生创新创业训练计划项目(202210132012)
CLC Number:
Cite this article
Luyao LI, Ning LYU, Yongjie ZHAI, Run ZHANG, Anxin YAN, Fuquan JIA. Genetic Polymorphism Analysis on Short Tandem Repeats of Y Chromosome in Han Population of Hohhot[J]. Current Biotechnology, 2025, 15(3): 544-554.
李璐瑶, 吕宁, 翟永结, 张润, 严安心, 贾富全. 呼和浩特汉族人群Y-STR的遗传多态性分析[J]. 生物技术进展, 2025, 15(3): 544-554.
share this article
| DYS449 | DYS627 | DYS481 | DYS557 | DYS635 | |||||
|---|---|---|---|---|---|---|---|---|---|
| (GD=0.870 6) | (GD=0.851 6) | (GD=0.798 1) | (GD=0.786 2) | (GD=0.771 9) | |||||
| 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 |
| 24 | 0.003 7 | 16 | 0.025 7 | 20 | 0.011 0 | 12 | 0.003 7 | 17 | 0.003 7 |
| 27 | 0.066 2 | 17 | 0.033 1 | 21 | 0.040 4 | 13 | 0.047 8 | 18 | 0.007 4 |
| 28 | 0.110 3 | 18 | 0.128 7 | 22 | 0.147 1 | 14 | 0.327 2 | 19 | 0.077 2 |
| 29 | 0.091 9 | 19 | 0.091 9 | 23 | 0.286 8 | 15 | 0.250 0 | 20 | 0.286 8 |
| 30 | 0.114 0 | 20 | 0.161 8 | 24 | 0.286 8 | 16 | 0.150 7 | 21 | 0.327 2 |
| 31 | 0.147 1 | 21 | 0.239 0 | 25 | 0.095 6 | 17 | 0.136 0 | 22 | 0.165 4 |
| 32 | 0.191 2 | 22 | 0.180 1 | 26 | 0.080 9 | 18 | 0.055 1 | 23 | 0.080 9 |
| 33 | 0.180 1 | 23 | 0.084 6 | 27 | 0.029 4 | 19 | 0.022 1 | 24 | 0.040 4 |
| 34 | 0.055 1 | 24 | 0.040 4 | 28 | 0.022 1 | 20 | 0.007 4 | 25 | 0.011 0 |
| 35 | 0.029 4 | 25 | 0.014 7 | ||||||
| 36 | 0.011 0 | ||||||||
Table 1 Allele frequencies and gene diversities of DYS449,DYS627,DYS481,DYS557 and DYS635 in Hohhot Han population
| DYS449 | DYS627 | DYS481 | DYS557 | DYS635 | |||||
|---|---|---|---|---|---|---|---|---|---|
| (GD=0.870 6) | (GD=0.851 6) | (GD=0.798 1) | (GD=0.786 2) | (GD=0.771 9) | |||||
| 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 |
| 24 | 0.003 7 | 16 | 0.025 7 | 20 | 0.011 0 | 12 | 0.003 7 | 17 | 0.003 7 |
| 27 | 0.066 2 | 17 | 0.033 1 | 21 | 0.040 4 | 13 | 0.047 8 | 18 | 0.007 4 |
| 28 | 0.110 3 | 18 | 0.128 7 | 22 | 0.147 1 | 14 | 0.327 2 | 19 | 0.077 2 |
| 29 | 0.091 9 | 19 | 0.091 9 | 23 | 0.286 8 | 15 | 0.250 0 | 20 | 0.286 8 |
| 30 | 0.114 0 | 20 | 0.161 8 | 24 | 0.286 8 | 16 | 0.150 7 | 21 | 0.327 2 |
| 31 | 0.147 1 | 21 | 0.239 0 | 25 | 0.095 6 | 17 | 0.136 0 | 22 | 0.165 4 |
| 32 | 0.191 2 | 22 | 0.180 1 | 26 | 0.080 9 | 18 | 0.055 1 | 23 | 0.080 9 |
| 33 | 0.180 1 | 23 | 0.084 6 | 27 | 0.029 4 | 19 | 0.022 1 | 24 | 0.040 4 |
| 34 | 0.055 1 | 24 | 0.040 4 | 28 | 0.022 1 | 20 | 0.007 4 | 25 | 0.011 0 |
| 35 | 0.029 4 | 25 | 0.014 7 | ||||||
| 36 | 0.011 0 | ||||||||
| DYS570 | DYS447 | DYS389II | DYS576 | DYS635 | |||||
|---|---|---|---|---|---|---|---|---|---|
| (GD=0.813 7) | (GD=0.789 7) | (GD=0.741 9) | (GD=0.786 1) | (GD=0.771 9) | |||||
| 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 |
| 14 | 0.014 7 | 22 | 0.014 7 | 26 | 0.007 4 | 15 | 0.022 1 | 17 | 0.003 7 |
| 15 | 0.051 5 | 23 | 0.272 1 | 27 | 0.106 6 | 16 | 0.088 2 | 18 | 0.007 4 |
| 16 | 0.176 5 | 24 | 0.279 4 | 28 | 0.371 3 | 17 | 0.209 6 | 19 | 0.077 2 |
| 17 | 0.250 0 | 25 | 0.183 8 | 29 | 0.286 8 | 18 | 0.330 9 | 20 | 0.286 8 |
| 18 | 0.235 3 | 26 | 0.147 1 | 30 | 0.161 8 | 19 | 0.205 9 | 21 | 0.327 2 |
| 19 | 0.183 8 | 27 | 0.069 9 | 31 | 0.055 1 | 20 | 0.106 6 | 22 | 0.165 4 |
| 20 | 0.044 1 | 28 | 0.018 4 | 32 | 0.011 0 | 21 | 0.036 8 | 23 | 0.080 9 |
| 21 | 0.040 4 | 29 | 0.014 7 | 24 | 0.040 4 | ||||
| 22 | 0.003 7 | 25 | 0.011 0 | ||||||
Table 2 Allele frequencies and gene diversities of DYS570, DYS447, DYS389II, DYS576 and DYS635 in Hohhot Han population
| DYS570 | DYS447 | DYS389II | DYS576 | DYS635 | |||||
|---|---|---|---|---|---|---|---|---|---|
| (GD=0.813 7) | (GD=0.789 7) | (GD=0.741 9) | (GD=0.786 1) | (GD=0.771 9) | |||||
| 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 |
| 14 | 0.014 7 | 22 | 0.014 7 | 26 | 0.007 4 | 15 | 0.022 1 | 17 | 0.003 7 |
| 15 | 0.051 5 | 23 | 0.272 1 | 27 | 0.106 6 | 16 | 0.088 2 | 18 | 0.007 4 |
| 16 | 0.176 5 | 24 | 0.279 4 | 28 | 0.371 3 | 17 | 0.209 6 | 19 | 0.077 2 |
| 17 | 0.250 0 | 25 | 0.183 8 | 29 | 0.286 8 | 18 | 0.330 9 | 20 | 0.286 8 |
| 18 | 0.235 3 | 26 | 0.147 1 | 30 | 0.161 8 | 19 | 0.205 9 | 21 | 0.327 2 |
| 19 | 0.183 8 | 27 | 0.069 9 | 31 | 0.055 1 | 20 | 0.106 6 | 22 | 0.165 4 |
| 20 | 0.044 1 | 28 | 0.018 4 | 32 | 0.011 0 | 21 | 0.036 8 | 23 | 0.080 9 |
| 21 | 0.040 4 | 29 | 0.014 7 | 24 | 0.040 4 | ||||
| 22 | 0.003 7 | 25 | 0.011 0 | ||||||
| DYS448 | DYS390 | DYS643 | DYS393 | DYS444 | |||||
|---|---|---|---|---|---|---|---|---|---|
| (GD=0.722 3) | (GD=0.679 4) | (GD=0.709 6) | (GD=0.608 2) | (GD=0.700 6) | |||||
| 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 |
| 16 | 0.003 7 | 21 | 0.014 7 | 8 | 0.018 4 | 10 | 0.003 7 | 10 | 0.007 4 |
| 17 | 0.029 4 | 22 | 0.044 1 | 9 | 0.095 6 | 12 | 0.558 8 | 11 | 0.110 3 |
| 18 | 0.117 6 | 23 | 0.477 9 | 10 | 0.312 5 | 13 | 0.239 0 | 12 | 0.371 3 |
| 19 | 0.393 4 | 24 | 0.239 0 | 11 | 0.408 1 | 14 | 0.150 7 | 13 | 0.367 6 |
| 20 | 0.312 5 | 25 | 0.183 8 | 12 | 0.136 0 | 15 | 0.044 1 | 14 | 0.128 7 |
| 21 | 0.110 3 | 26 | 0.040 4 | 13 | 0.029 4 | 16 | 0.003 7 | 15 | 0.011 0 |
| 22 | 0.033 1 | 16 | 0.003 7 | ||||||
Table 3 Allele frequencies and gene diversities of DYS448,DYS390,DYS643,DYS393 and DYS444 in Hohhot Han population
| DYS448 | DYS390 | DYS643 | DYS393 | DYS444 | |||||
|---|---|---|---|---|---|---|---|---|---|
| (GD=0.722 3) | (GD=0.679 4) | (GD=0.709 6) | (GD=0.608 2) | (GD=0.700 6) | |||||
| 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 |
| 16 | 0.003 7 | 21 | 0.014 7 | 8 | 0.018 4 | 10 | 0.003 7 | 10 | 0.007 4 |
| 17 | 0.029 4 | 22 | 0.044 1 | 9 | 0.095 6 | 12 | 0.558 8 | 11 | 0.110 3 |
| 18 | 0.117 6 | 23 | 0.477 9 | 10 | 0.312 5 | 13 | 0.239 0 | 12 | 0.371 3 |
| 19 | 0.393 4 | 24 | 0.239 0 | 11 | 0.408 1 | 14 | 0.150 7 | 13 | 0.367 6 |
| 20 | 0.312 5 | 25 | 0.183 8 | 12 | 0.136 0 | 15 | 0.044 1 | 14 | 0.128 7 |
| 21 | 0.110 3 | 26 | 0.040 4 | 13 | 0.029 4 | 16 | 0.003 7 | 15 | 0.011 0 |
| 22 | 0.033 1 | 16 | 0.003 7 | ||||||
| DYS19 | DYS456 | DYS438 | DYS391 | DYS460 | |||||
|---|---|---|---|---|---|---|---|---|---|
| (GD=0.706 4) | (GD=0.608 6) | (GD=0.491 8) | (GD=0.377 5) | (GD=0.667 5) | |||||
| 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 |
| 13 | 0.033 1 | 13 | 0.018 4 | 9 | 0.033 1 | 7 | 0.014 7 | 8 | 0.003 7 |
| 14 | 0.246 3 | 14 | 0.147 1 | 10 | 0.661 8 | 9 | 0.025 7 | 9 | 0.352 9 |
| 15 | 0.422 8 | 15 | 0.569 9 | 11 | 0.264 7 | 10 | 0.768 4 | 10 | 0.426 5 |
| 16 | 0.224 3 | 16 | 0.209 6 | 12 | 0.025 7 | 11 | 0.180 1 | 11 | 0.158 1 |
| 17 | 0.073 5 | 17 | 0.055 1 | 13 | 0.014 7 | 12 | 0.011 0 | 12 | 0.058 8 |
Table 4 Allele frequencies and gene diversities of DYS19, DYS456, DYS438, DYS391 and DYS460 in Hohhot Han population
| DYS19 | DYS456 | DYS438 | DYS391 | DYS460 | |||||
|---|---|---|---|---|---|---|---|---|---|
| (GD=0.706 4) | (GD=0.608 6) | (GD=0.491 8) | (GD=0.377 5) | (GD=0.667 5) | |||||
| 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 |
| 13 | 0.033 1 | 13 | 0.018 4 | 9 | 0.033 1 | 7 | 0.014 7 | 8 | 0.003 7 |
| 14 | 0.246 3 | 14 | 0.147 1 | 10 | 0.661 8 | 9 | 0.025 7 | 9 | 0.352 9 |
| 15 | 0.422 8 | 15 | 0.569 9 | 11 | 0.264 7 | 10 | 0.768 4 | 10 | 0.426 5 |
| 16 | 0.224 3 | 16 | 0.209 6 | 12 | 0.025 7 | 11 | 0.180 1 | 11 | 0.158 1 |
| 17 | 0.073 5 | 17 | 0.055 1 | 13 | 0.014 7 | 12 | 0.011 0 | 12 | 0.058 8 |
| DYS596 | DYS522 | DYS549 | YGATAH4 | DYS439 | |||||
|---|---|---|---|---|---|---|---|---|---|
| (GD=0.611 0) | (GD=0.677 5) | (GD=0.588 5) | (GD=0.547 2) | (GD=0.703 2) | |||||
| 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 |
| 13 | 0.011 0 | 9 | 0.011 0 | 10 | 0.014 7 | 10 | 0.047 8 | 10 | 0.088 2 |
| 14 | 0.393 4 | 10 | 0.099 3 | 11 | 0.025 7 | 11 | 0.268 4 | 11 | 0.349 3 |
| 15 | 0.474 3 | 11 | 0.319 9 | 12 | 0.533 1 | 12 | 0.614 0 | 12 | 0.382 4 |
| 16 | 0.106 6 | 12 | 0.452 2 | 13 | 0.352 9 | 13 | 0.058 8 | 13 | 0.150 7 |
| 17 | 0.007 4 | 13 | 0.084 6 | 14 | 0.062 5 | 14 | 0.007 4 | 14 | 0.025 7 |
| 18 | 0.007 4 | 14 | 0.033 1 | 15 | 0.011 0 | 15 | 0.003 7 | 15 | 0.003 7 |
Table 5 Allele frequencies and gene diversities of DYS596, DYS522, DYS549, YGATAH4 and DYS439 in Hohhot Han population
| DYS596 | DYS522 | DYS549 | YGATAH4 | DYS439 | |||||
|---|---|---|---|---|---|---|---|---|---|
| (GD=0.611 0) | (GD=0.677 5) | (GD=0.588 5) | (GD=0.547 2) | (GD=0.703 2) | |||||
| 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 |
| 13 | 0.011 0 | 9 | 0.011 0 | 10 | 0.014 7 | 10 | 0.047 8 | 10 | 0.088 2 |
| 14 | 0.393 4 | 10 | 0.099 3 | 11 | 0.025 7 | 11 | 0.268 4 | 11 | 0.349 3 |
| 15 | 0.474 3 | 11 | 0.319 9 | 12 | 0.533 1 | 12 | 0.614 0 | 12 | 0.382 4 |
| 16 | 0.106 6 | 12 | 0.452 2 | 13 | 0.352 9 | 13 | 0.058 8 | 13 | 0.150 7 |
| 17 | 0.007 4 | 13 | 0.084 6 | 14 | 0.062 5 | 14 | 0.007 4 | 14 | 0.025 7 |
| 18 | 0.007 4 | 14 | 0.033 1 | 15 | 0.011 0 | 15 | 0.003 7 | 15 | 0.003 7 |
| DYS533 | DYS593 | DYS389I | DYS437 | DYS645 | |||||
|---|---|---|---|---|---|---|---|---|---|
| (GD=0.554 1) | (GD=0.602 5) | (GD=0.589 7) | (GD=0.529 3) | (GD=0.043 4) | |||||
| 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 |
| 10 | 0.044 1 | 14 | 0.003 7 | 11 | 0.011 0 | 13 | 0.025 7 | 6.4 | 0.003 7 |
| 11 | 0.584 6 | 15 | 0.231 6 | 12 | 0.540 4 | 14 | 0.555 1 | 8 | 0.977 9 |
| 12 | 0.319 9 | 16 | 0.562 5 | 13 | 0.323 5 | 15 | 0.404 4 | 9 | 0.018 4 |
| 13 | 0.044 1 | 17 | 0.169 1 | 14 | 0.125 0 | 16 | 0.014 7 | ||
| 14 | 0.007 4 | 18 | 0.033 1 | ||||||
Table 6 Allele frequencies and gene diversities of DYS533, DYS593, DYS389I, DYS437 and DYS645 in Hohhot Han population
| DYS533 | DYS593 | DYS389I | DYS437 | DYS645 | |||||
|---|---|---|---|---|---|---|---|---|---|
| (GD=0.554 1) | (GD=0.602 5) | (GD=0.589 7) | (GD=0.529 3) | (GD=0.043 4) | |||||
| 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 |
| 10 | 0.044 1 | 14 | 0.003 7 | 11 | 0.011 0 | 13 | 0.025 7 | 6.4 | 0.003 7 |
| 11 | 0.584 6 | 15 | 0.231 6 | 12 | 0.540 4 | 14 | 0.555 1 | 8 | 0.977 9 |
| 12 | 0.319 9 | 16 | 0.562 5 | 13 | 0.323 5 | 15 | 0.404 4 | 9 | 0.018 4 |
| 13 | 0.044 1 | 17 | 0.169 1 | 14 | 0.125 0 | 16 | 0.014 7 | ||
| 14 | 0.007 4 | 18 | 0.033 1 | ||||||
| DYS527 | DYS518 | DYS458 | |||||
|---|---|---|---|---|---|---|---|
| (GD=0.943 0) | (GD=0.858 9) | (GD=0.820 7) | |||||
| 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 |
| 17,21 | 0.003 7 | 20,25 | 0.003 7 | 32 | 0.003 7 | 13 | 0.003 7 |
| 18,19 | 0.003 7 | 21,21 | 0.069 9 | 33 | 0.029 4 | 14 | 0.014 7 |
| 18,20 | 0.003 7 | 21,22 | 0.084 6 | 34 | 0.011 0 | 15 | 0.095 6 |
| 18,23 | 0.003 7 | 21,23 | 0.102 9 | 35 | 0.106 6 | 16 | 0.187 5 |
| 19,19 | 0.003 7 | 21,24 | 0.040 4 | 36 | 0.202 2 | 17 | 0.235 3 |
| 19,20 | 0.051 5 | 22,22 | 0.066 2 | 36.2 | 0.003 7 | 17.2 | 0.003 7 |
| 19,21 | 0.025 7 | 22,23 | 0.066 2 | 37 | 0.180 1 | 18 | 0.227 9 |
| 19,22 | 0.007 4 | 22,24 | 0.047 8 | 37.2 | 0.007 4 | 19 | 0.165 4 |
| 19,23 | 0.014 7 | 22,25 | 0.011 0 | 38 | 0.176 5 | 19.2 | 0.003 7 |
| 20,20 | 0.025 7 | 23,23 | 0.058 8 | 39 | 0.147 1 | 20 | 0.055 1 |
| 20,21 | 0.018 4 | 23,24 | 0.018 4 | 40 | 0.055 1 | 21 | 0.003 7 |
| 20,22 | 0.044 1 | 24,24 | 0.040 4 | 41 | 0.044 1 | 22 | 0.003 7 |
| 20,23 | 0.088 2 | 24,25 | 0.014 7 | 42 | 0.025 7 | ||
| 20,24 | 0.069 9 | 25,25 | 0.011 0 | 43 | 0.007 4 | ||
Table 7 Allele frequencies and gene diversities of DYS527, DYS518 and DYS458 in Hohhot Han population
| DYS527 | DYS518 | DYS458 | |||||
|---|---|---|---|---|---|---|---|
| (GD=0.943 0) | (GD=0.858 9) | (GD=0.820 7) | |||||
| 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 |
| 17,21 | 0.003 7 | 20,25 | 0.003 7 | 32 | 0.003 7 | 13 | 0.003 7 |
| 18,19 | 0.003 7 | 21,21 | 0.069 9 | 33 | 0.029 4 | 14 | 0.014 7 |
| 18,20 | 0.003 7 | 21,22 | 0.084 6 | 34 | 0.011 0 | 15 | 0.095 6 |
| 18,23 | 0.003 7 | 21,23 | 0.102 9 | 35 | 0.106 6 | 16 | 0.187 5 |
| 19,19 | 0.003 7 | 21,24 | 0.040 4 | 36 | 0.202 2 | 17 | 0.235 3 |
| 19,20 | 0.051 5 | 22,22 | 0.066 2 | 36.2 | 0.003 7 | 17.2 | 0.003 7 |
| 19,21 | 0.025 7 | 22,23 | 0.066 2 | 37 | 0.180 1 | 18 | 0.227 9 |
| 19,22 | 0.007 4 | 22,24 | 0.047 8 | 37.2 | 0.007 4 | 19 | 0.165 4 |
| 19,23 | 0.014 7 | 22,25 | 0.011 0 | 38 | 0.176 5 | 19.2 | 0.003 7 |
| 20,20 | 0.025 7 | 23,23 | 0.058 8 | 39 | 0.147 1 | 20 | 0.055 1 |
| 20,21 | 0.018 4 | 23,24 | 0.018 4 | 40 | 0.055 1 | 21 | 0.003 7 |
| 20,22 | 0.044 1 | 24,24 | 0.040 4 | 41 | 0.044 1 | 22 | 0.003 7 |
| 20,23 | 0.088 2 | 24,25 | 0.014 7 | 42 | 0.025 7 | ||
| 20,24 | 0.069 9 | 25,25 | 0.011 0 | 43 | 0.007 4 | ||
| DYS385 | DYF387S1 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| (GD=0.967 5) | (GD=0.946 7) | ||||||||
| 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 |
| 9,16 | 0.003 7 | 12,14 | 0.011 0 | 13,22 | 0.014 7 | 33,37 | 0.007 4 | 36,40 | 0.022 1 |
| 10,16 | 0.011 0 | 12,15 | 0.014 7 | 13,23 | 0.003 7 | 34,34 | 0.007 4 | 36,41 | 0.003 7 |
| 10,17 | 0.003 7 | 12,16 | 0.058 8 | 14,14 | 0.003 7 | 34,36 | 0.003 7 | 37,37 | 0.058 8 |
| 10,18 | 0.003 7 | 12,17 | 0.018 4 | 14,15 | 0.007 4 | 34,37 | 0.007 4 | 37,38 | 0.077 2 |
| 10,19 | 0.011 0 | 12,18 | 0.055 1 | 14,16 | 0.018 4 | 34,38 | 0.011 0 | 37,39 | 0.036 8 |
| 11,11 | 0.033 1 | 12,19 | 0.058 8 | 14,17 | 0.011 0 | 34,40 | 0.003 7 | 37,40 | 0.018 4 |
| 11,12 | 0.029 4 | 12,20 | 0.011 0 | 14,18 | 0.047 8 | 35,35 | 0.018 4 | 37,42 | 0.003 7 |
| 11,13 | 0.025 7 | 12,22 | 0.003 7 | 14,19 | 0.007 4 | 35,36 | 0.011 0 | 38,38 | 0.073 5 |
| 11,14 | 0.025 7 | 13,13 | 0.047 8 | 14,20 | 0.003 7 | 35,37 | 0.033 1 | 38,39 | 0.058 8 |
| 11,15 | 0.011 0 | 13,14 | 0.007 4 | 15,16 | 0.003 7 | 35,38 | 0.080 9 | 38,40 | 0.022 1 |
| 11,16 | 0.022 1 | 13,15 | 0.007 4 | 15,17 | 0.018 4 | 35,39 | 0.077 2 | 39,39 | 0.022 1 |
| 11,17 | 0.014 7 | 13,17 | 0.036 8 | 15,18 | 0.007 4 | 35,40 | 0.022 1 | 39,40 | 0.007 4 |
| 11,18 | 0.022 1 | 13,18 | 0.058 8 | 15,21 | 0.007 4 | 35,41 | 0.014 7 | 39,41 | 0.007 4 |
| 11,19 | 0.040 4 | 13,19 | 0.066 2 | 18,19 | 0.007 4 | 36,36 | 0.058 8 | 40,40 | 0.018 4 |
| 11,20 | 0.014 7 | 13,20 | 0.029 4 | 19,19 | 0.011 0 | 36,37 | 0.025 7 | 40,41 | 0.007 4 |
| 12,12 | 0.044 1 | 13,21 | 0.003 7 | 19,20 | 0.003 7 | 36,38 | 0.091 9 | 41,41 | 0.003 7 |
| 12,13 | 0.018 4 | 36,39 | 0.084 6 | ||||||
Table 8 Allele frequencies and gene diversities of DYS385 and DYF387S1 in Hohhot Han population
| DYS385 | DYF387S1 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| (GD=0.967 5) | (GD=0.946 7) | ||||||||
| 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 | 等位基因 | 基因频率 |
| 9,16 | 0.003 7 | 12,14 | 0.011 0 | 13,22 | 0.014 7 | 33,37 | 0.007 4 | 36,40 | 0.022 1 |
| 10,16 | 0.011 0 | 12,15 | 0.014 7 | 13,23 | 0.003 7 | 34,34 | 0.007 4 | 36,41 | 0.003 7 |
| 10,17 | 0.003 7 | 12,16 | 0.058 8 | 14,14 | 0.003 7 | 34,36 | 0.003 7 | 37,37 | 0.058 8 |
| 10,18 | 0.003 7 | 12,17 | 0.018 4 | 14,15 | 0.007 4 | 34,37 | 0.007 4 | 37,38 | 0.077 2 |
| 10,19 | 0.011 0 | 12,18 | 0.055 1 | 14,16 | 0.018 4 | 34,38 | 0.011 0 | 37,39 | 0.036 8 |
| 11,11 | 0.033 1 | 12,19 | 0.058 8 | 14,17 | 0.011 0 | 34,40 | 0.003 7 | 37,40 | 0.018 4 |
| 11,12 | 0.029 4 | 12,20 | 0.011 0 | 14,18 | 0.047 8 | 35,35 | 0.018 4 | 37,42 | 0.003 7 |
| 11,13 | 0.025 7 | 12,22 | 0.003 7 | 14,19 | 0.007 4 | 35,36 | 0.011 0 | 38,38 | 0.073 5 |
| 11,14 | 0.025 7 | 13,13 | 0.047 8 | 14,20 | 0.003 7 | 35,37 | 0.033 1 | 38,39 | 0.058 8 |
| 11,15 | 0.011 0 | 13,14 | 0.007 4 | 15,16 | 0.003 7 | 35,38 | 0.080 9 | 38,40 | 0.022 1 |
| 11,16 | 0.022 1 | 13,15 | 0.007 4 | 15,17 | 0.018 4 | 35,39 | 0.077 2 | 39,39 | 0.022 1 |
| 11,17 | 0.014 7 | 13,17 | 0.036 8 | 15,18 | 0.007 4 | 35,40 | 0.022 1 | 39,40 | 0.007 4 |
| 11,18 | 0.022 1 | 13,18 | 0.058 8 | 15,21 | 0.007 4 | 35,41 | 0.014 7 | 39,41 | 0.007 4 |
| 11,19 | 0.040 4 | 13,19 | 0.066 2 | 18,19 | 0.007 4 | 36,36 | 0.058 8 | 40,40 | 0.018 4 |
| 11,20 | 0.014 7 | 13,20 | 0.029 4 | 19,19 | 0.011 0 | 36,37 | 0.025 7 | 40,41 | 0.007 4 |
| 12,12 | 0.044 1 | 13,21 | 0.003 7 | 19,20 | 0.003 7 | 36,38 | 0.091 9 | 41,41 | 0.003 7 |
| 12,13 | 0.018 4 | 36,39 | 0.084 6 | ||||||
| 群体 | 呼和浩特汉族 | 贵州布依族 | 云南傣族 | 湖南侗族 | 四川客家汉族 | 陕西汉族 | 四川汉族 | 湖南苗族 | 呼伦贝尔蒙古族 | 鄂尔多斯蒙古族 | 新疆蒙古族 | 北川羌族 | 青海撒拉族 | 成都藏族 | 甘孜藏族 | 拉萨藏族 | 那曲藏族 | 阿里藏族 | 青海藏族 | 日喀则藏族 | 湖南土家族 | 贵阳彝族 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 呼和浩特汉族 | - | |||||||||||||||||||||
| 贵州布依族 | 0.052 8 | - | ||||||||||||||||||||
| 云南傣族 | 0.074 3 | 0.062 5 | - | |||||||||||||||||||
| 湖南侗族 | 0.047 7 | 0.054 3 | 0.083 6 | - | ||||||||||||||||||
| 四川客家汉族 | 0.015 8 | 0.025 3 | 0.072 3 | 0.056 | - | |||||||||||||||||
| 陕西汉族 | 0.001 4 | 0.058 2 | 0.080 9 | 0.045 1 | 0.022 0 | - | ||||||||||||||||
| 四川汉族 | 0.011 9 | 0.028 9 | 0.067 3 | 0.025 9 | 0.010 7 | 0.010 9 | - | |||||||||||||||
| 湖南苗族 | 0.011 1 | 0.036 8 | 0.077 2 | 0.016 4 | 0.017 6 | 0.009 6 | 0.003 9 | - | ||||||||||||||
| 呼伦贝尔蒙古族 | 0.101 0 | 0.122 5 | 0.159 3 | 0.045 1 | 0.125 6 | 0.096 1 | 0.083 7 | 0.061 0 | - | |||||||||||||
| 鄂尔多斯蒙古族 | 0.054 0 | 0.099 9 | 0.140 5 | 0.031 4 | 0.082 0 | 0.043 7 | 0.048 2 | 0.027 9 | 0.016 7 | - | ||||||||||||
| 新疆蒙古族 | 0.089 8 | 0.101 1 | 0.166 5 | 0.040 5 | 0.106 6 | 0.081 7 | 0.067 6 | 0.046 8 | 0.017 3 | 0.013 5 | - | |||||||||||
| 北川羌族 | 0.004 2 | 0.032 8 | 0.072 4 | 0.044 6 | 0.003 4 | 0.007 5 | 0.004 2 | 0.009 0 | 0.107 6 | 0.063 1 | 0.089 5 | - | ||||||||||
| 青海撒拉族 | 0.092 4 | 0.081 2 | 0.145 4 | 0.060 4 | 0.104 3 | 0.084 3 | 0.075 7 | 0.057 7 | 0.097 5 | 0.061 1 | 0.041 9 | 0.085 7 | - | |||||||||
| 成都藏族 | 0.131 7 | 0.112 3 | 0.194 5 | 0.113 9 | 0.126 3 | 0.125 1 | 0.084 1 | 0.093 8 | 0.132 7 | 0.112 0 | 0.090 6 | 0.107 8 | 0.127 6 | - | ||||||||
| 甘孜藏族 | 0.311 7 | 0.323 8 | 0.389 7 | 0.254 7 | 0.333 6 | 0.294 2 | 0.264 5 | 0.249 9 | 0.242 3 | 0.215 7 | 0.190 0 | 0.295 9 | 0.257 7 | 0.137 1 | - | |||||||
| 拉萨藏族 | 0.086 2 | 0.132 8 | 0.179 5 | 0.093 7 | 0.111 3 | 0.075 2 | 0.067 9 | 0.066 7 | 0.114 9 | 0.065 6 | 0.071 3 | 0.081 5 | 0.090 8 | 0.039 9 | 0.112 7 | - | ||||||
| 那曲藏族 | 0.124 6 | 0.167 5 | 0.230 0 | 0.109 7 | 0.144 4 | 0.110 4 | 0.098 6 | 0.090 9 | 0.116 5 | 0.069 9 | 0.072 1 | 0.117 1 | 0.108 4 | 0.050 5 | 0.068 8 | 0.003 4 | - | |||||
| 阿里藏族 | 0.077 7 | 0.128 9 | 0.170 0 | 0.097 2 | 0.104 7 | 0.067 5 | 0.063 8 | 0.066 3 | 0.127 6 | 0.072 2 | 0.079 6 | 0.075 0 | 0.085 3 | 0.045 4 | 0.127 7 | 0.014 6 | 0.003 7 | - | ||||
| 青海藏族 | 0.210 1 | 0.238 4 | 0.296 8 | 0.171 7 | 0.230 3 | 0.194 2 | 0.175 3 | 0.162 8 | 0.155 1 | 0.125 8 | 0.107 8 | 0.200 5 | 0.167 2 | 0.080 5 | 0.014 3 | 0.041 8 | 0.017 9 | 0.054 2 | - | |||
| 日喀则藏族 | 0.068 8 | 0.114 6 | 0.158 0 | 0.086 9 | 0.086 9 | 0.059 7 | 0.051 3 | 0.054 8 | 0.105 7 | 0.060 9 | 0.069 9 | 0.063 0 | 0.095 9 | 0.031 6 | 0.127 0 | 0.013 1 | 0.004 5 | 0.011 4 | 0.052 2 | - | ||
| 湖南土家族 | 0.010 9 | 0.037 7 | 0.083 1 | 0.020 9 | 0.015 2 | 0.010 1 | 0.003 9 | 0.000 1 | 0.072 7 | 0.034 7 | 0.054 4 | 0.008 0 | 0.065 6 | 0.097 8 | 0.257 4 | 0.069 9 | 0.095 4 | 0.068 2 | 0.169 0 | 0.057 5 | - | |
| 贵阳彝族 | 0.027 7 | 0.037 6 | 0.070 9 | 0.025 4 | 0.025 4 | 0.019 3 | 0.008 6 | 0.012 7 | 0.087 5 | 0.047 8 | 0.069 7 | 0.018 7 | 0.063 3 | 0.110 8 | 0.276 4 | 0.084 5 | 0.108 4 | 0.078 5 | 0.187 9 | 0.071 9 | 0.014 5 | - |
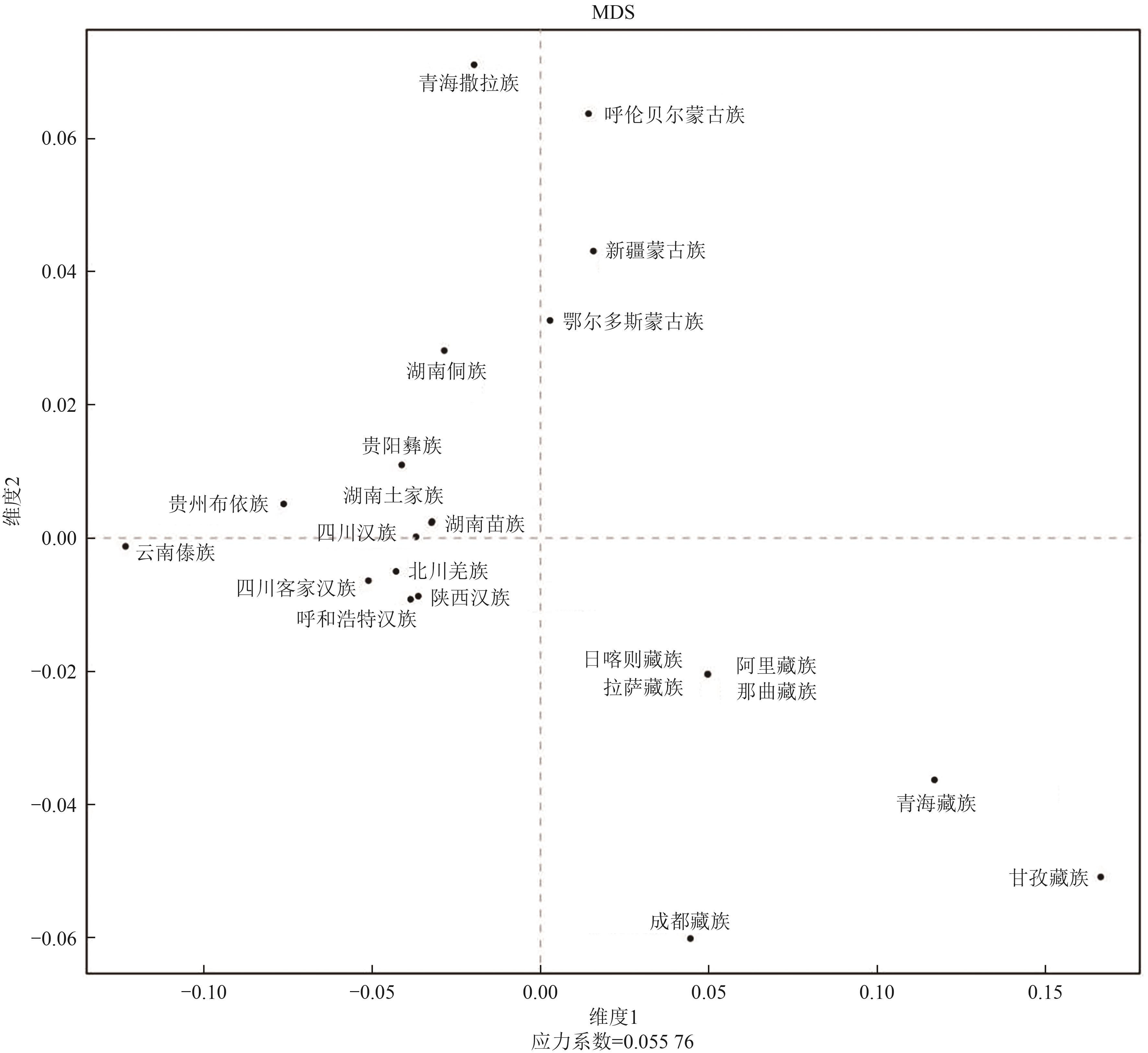
Table 9 Pairwise genetic distance (Rst) between Hohhot Han and other 21 Chinese populations
| 群体 | 呼和浩特汉族 | 贵州布依族 | 云南傣族 | 湖南侗族 | 四川客家汉族 | 陕西汉族 | 四川汉族 | 湖南苗族 | 呼伦贝尔蒙古族 | 鄂尔多斯蒙古族 | 新疆蒙古族 | 北川羌族 | 青海撒拉族 | 成都藏族 | 甘孜藏族 | 拉萨藏族 | 那曲藏族 | 阿里藏族 | 青海藏族 | 日喀则藏族 | 湖南土家族 | 贵阳彝族 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 呼和浩特汉族 | - | |||||||||||||||||||||
| 贵州布依族 | 0.052 8 | - | ||||||||||||||||||||
| 云南傣族 | 0.074 3 | 0.062 5 | - | |||||||||||||||||||
| 湖南侗族 | 0.047 7 | 0.054 3 | 0.083 6 | - | ||||||||||||||||||
| 四川客家汉族 | 0.015 8 | 0.025 3 | 0.072 3 | 0.056 | - | |||||||||||||||||
| 陕西汉族 | 0.001 4 | 0.058 2 | 0.080 9 | 0.045 1 | 0.022 0 | - | ||||||||||||||||
| 四川汉族 | 0.011 9 | 0.028 9 | 0.067 3 | 0.025 9 | 0.010 7 | 0.010 9 | - | |||||||||||||||
| 湖南苗族 | 0.011 1 | 0.036 8 | 0.077 2 | 0.016 4 | 0.017 6 | 0.009 6 | 0.003 9 | - | ||||||||||||||
| 呼伦贝尔蒙古族 | 0.101 0 | 0.122 5 | 0.159 3 | 0.045 1 | 0.125 6 | 0.096 1 | 0.083 7 | 0.061 0 | - | |||||||||||||
| 鄂尔多斯蒙古族 | 0.054 0 | 0.099 9 | 0.140 5 | 0.031 4 | 0.082 0 | 0.043 7 | 0.048 2 | 0.027 9 | 0.016 7 | - | ||||||||||||
| 新疆蒙古族 | 0.089 8 | 0.101 1 | 0.166 5 | 0.040 5 | 0.106 6 | 0.081 7 | 0.067 6 | 0.046 8 | 0.017 3 | 0.013 5 | - | |||||||||||
| 北川羌族 | 0.004 2 | 0.032 8 | 0.072 4 | 0.044 6 | 0.003 4 | 0.007 5 | 0.004 2 | 0.009 0 | 0.107 6 | 0.063 1 | 0.089 5 | - | ||||||||||
| 青海撒拉族 | 0.092 4 | 0.081 2 | 0.145 4 | 0.060 4 | 0.104 3 | 0.084 3 | 0.075 7 | 0.057 7 | 0.097 5 | 0.061 1 | 0.041 9 | 0.085 7 | - | |||||||||
| 成都藏族 | 0.131 7 | 0.112 3 | 0.194 5 | 0.113 9 | 0.126 3 | 0.125 1 | 0.084 1 | 0.093 8 | 0.132 7 | 0.112 0 | 0.090 6 | 0.107 8 | 0.127 6 | - | ||||||||
| 甘孜藏族 | 0.311 7 | 0.323 8 | 0.389 7 | 0.254 7 | 0.333 6 | 0.294 2 | 0.264 5 | 0.249 9 | 0.242 3 | 0.215 7 | 0.190 0 | 0.295 9 | 0.257 7 | 0.137 1 | - | |||||||
| 拉萨藏族 | 0.086 2 | 0.132 8 | 0.179 5 | 0.093 7 | 0.111 3 | 0.075 2 | 0.067 9 | 0.066 7 | 0.114 9 | 0.065 6 | 0.071 3 | 0.081 5 | 0.090 8 | 0.039 9 | 0.112 7 | - | ||||||
| 那曲藏族 | 0.124 6 | 0.167 5 | 0.230 0 | 0.109 7 | 0.144 4 | 0.110 4 | 0.098 6 | 0.090 9 | 0.116 5 | 0.069 9 | 0.072 1 | 0.117 1 | 0.108 4 | 0.050 5 | 0.068 8 | 0.003 4 | - | |||||
| 阿里藏族 | 0.077 7 | 0.128 9 | 0.170 0 | 0.097 2 | 0.104 7 | 0.067 5 | 0.063 8 | 0.066 3 | 0.127 6 | 0.072 2 | 0.079 6 | 0.075 0 | 0.085 3 | 0.045 4 | 0.127 7 | 0.014 6 | 0.003 7 | - | ||||
| 青海藏族 | 0.210 1 | 0.238 4 | 0.296 8 | 0.171 7 | 0.230 3 | 0.194 2 | 0.175 3 | 0.162 8 | 0.155 1 | 0.125 8 | 0.107 8 | 0.200 5 | 0.167 2 | 0.080 5 | 0.014 3 | 0.041 8 | 0.017 9 | 0.054 2 | - | |||
| 日喀则藏族 | 0.068 8 | 0.114 6 | 0.158 0 | 0.086 9 | 0.086 9 | 0.059 7 | 0.051 3 | 0.054 8 | 0.105 7 | 0.060 9 | 0.069 9 | 0.063 0 | 0.095 9 | 0.031 6 | 0.127 0 | 0.013 1 | 0.004 5 | 0.011 4 | 0.052 2 | - | ||
| 湖南土家族 | 0.010 9 | 0.037 7 | 0.083 1 | 0.020 9 | 0.015 2 | 0.010 1 | 0.003 9 | 0.000 1 | 0.072 7 | 0.034 7 | 0.054 4 | 0.008 0 | 0.065 6 | 0.097 8 | 0.257 4 | 0.069 9 | 0.095 4 | 0.068 2 | 0.169 0 | 0.057 5 | - | |
| 贵阳彝族 | 0.027 7 | 0.037 6 | 0.070 9 | 0.025 4 | 0.025 4 | 0.019 3 | 0.008 6 | 0.012 7 | 0.087 5 | 0.047 8 | 0.069 7 | 0.018 7 | 0.063 3 | 0.110 8 | 0.276 4 | 0.084 5 | 0.108 4 | 0.078 5 | 0.187 9 | 0.071 9 | 0.014 5 | - |
| 1 | RALF A, LUBACH D, KOUSOURI N, et al.. Identification and characterization of novel rapidly mutating Y-chromosomal short tandem repeat markers[J]. Hum. Mutat., 2020, 41(9): 1680-1696. |
| 2 | JOBLING M A, TYLER-SMITH C. Human Y-chromosome variation in the genome-sequencing era[J]. Nat. Rev. Genet., 2017, 18(8): 485-497. |
| 3 | PURPS J, GEPPERT M, NAGY M, et al.. Validation of a combined autosomal/Y-chromosomal STR approach for analyzing typical biological stains in sexual-assault cases[J]. Forensic Sci. Int. Genet., 2015, 19: 238-242. |
| 4 | SONG M, SONG F, WANG S, et al.. Developmental validation of the Yfiler platinum PCR amplification kit for forensic genetic caseworks and databases[J]. Electrophoresis, 2021, 42(1-2): 126-133. |
| 5 | ZHAO Y, YAO X, LI Y, et al.. Developmental validation of the microreaderTM Y prime plus ID system: an advanced Y-STR 38-plex system for forensic applications[J]. Sci. Justice, 2021, 61(3): 260-270. |
| 6 | 孙宽,侯一平.法医族源推断的分子生物学进展[J].法医学杂志,2018,34(3):286-293. |
| SUN K, HOU Y P. Progress on molecular biology for forensic ancestry inference[J]. J. Forensic Med., 2018, 34(3): 286-293. | |
| 7 | LI L, YAO L, GONG H, et al.. Genetic characterisation for Yan'an Han population in northern Shaanxi Province, China, via 38Y-STRs using yfilerTM platinum[J]. Ann. Hum. Biol., 2021, 48(4): 327-334. |
| 8 | NEI M. Analysis of gene diversity in subdivided populations[J]. Proc. Natl. Acad. Sci. USA, 1973, 70(12): 3321-3323. |
| 9 | WILLUWEIT S, ROEWER L. The new Y chromosome haplotype reference database[J]. Forensic Sci. Int. Genet., 2015, 15: 43-48. |
| 10 | TYTGAT O, ŠKEVIN S, DEFORCE D, et al.. Nanopore sequencing of a forensic combined STR and SNP multiplex[J/OL]. Forensic Sci. Int. Genet., 2022, 56: 102621[2025-02-20]. . |
| 11 | YANG C, LIU C, LUN M, et al.. Dissecting the genetic admixture and forensic signatures of ethnolinguistically diverse Chinese populations via a 114-plex NGS InDel panel[J/OL]. BMC Genomics, 2024, 25(1): 1137[2025-02-20]. . |
| 12 | 李茜,魏天莉,程良红,等.中国南方汉族非亲缘男性个体Y-STR基因座的遗传多态性[J].中国输血杂志,2009,22(5):387-391. |
| LI Q, WEI T L, CHENG L H, et al.. Genetic polymorphism of Y-STR loci in unrelated male individuals of Han nationality in Southern China[J]. Chin. J. Blood Transfus., 2009, 22(5): 387-391. | |
| 13 | VARGA T, KEYSER C, BEER Z, et al.. Short tandem repeat data analysis in a Mongolian population[J]. Leg. Med. (Tokyo), 2003, 5(): S156-S159. |
| 14 | 刘淑萍,苏秀兰,毕力夫,等.内蒙古农区蒙古族D3S1358、D13S317和D5S818基因座位的遗传多态性[J].中华医学遗传学杂志,2004,21(3):297-298. |
| LIU S P, SU X L, BI L F, et al.. Genetic polymorphisms of short tandem repeat loci D3S1358, D13S317, D5S818 from Nongqu Mongolia[J]. Chin. J. Med. Genet., 2004, 21(3): 297-298. | |
| 15 | SU X, LI D, LIFU S L B, et al.. Population studies on two native Mongolian population groups in China using STR loci[J]. Forensic Sci. Int., 2004, 141(2-3): 197-199. |
| 16 | YUAN Q Q, XIAO S Y, FAROUK O, et al.. Management of granulomatous lobular mastitis: an international multidisciplinary consensus (2021 edition)[J/OL]. Mil. Med. Res., 2022, 9(1): 20[2025-02-20]. . |
| 17 | JIN X, XING G, YANG C, et al.. Genetic polymorphisms of 44 Y chromosomal genetic markers in the Inner Mongolia Han population and its genetic relationship analysis with other reference populations[J]. Forensic Sci. Res., 2022, 7(3): 510-517. |
| 18 | LI M, ZHOU W, ZHANG Y, et al.. Development and validation of a novel 29-plex Y-STR typing system for forensic application[J/OL]. Forensic Sci. Int. Genet., 2020, 44: 102169[2025-02-20]. . |
| 19 | CHAI S, LI M, TAO R, et al.. Internal validation of an improved system for forensic application: a 41-plex Y-STR panel[J]. Forensic Sci. Res., 2023, 8(1): 70-78. |
| 20 | 韩月雯,胡宗利,陈国平,等.重庆汉族人群5个Y-STR基因座的遗传多态性分析[J].西安交通大学学报(医学版),2013,34(1):29-33. |
| HAN Y W, HU Z L, CHEN G P, et al.. Analysis of genetic polymorphisms of 5 Y-STR loci in the Han population in Chongqing[J]. J. Xi'an Jiaotong Univ. Med. Sci., 2013, 34(1): 29-33. |
| [1] | Dayu LIU, Jichao XU, Ying ZENG, Quyi XU, Hong LIU, Yue LI, Changhui LIU. Efficacy Evaluation of Forensic Polymorphism Application of 59 Y-STR [J]. Current Biotechnology, 2023, 13(1): 107-114. |
| [2] | Longfei MIAO, Dapeng SUN, Wenhua MA, Jianjun ZHONG. Analysis of Genetic Polymorphisms at 37 Y⁃STR Loci in Dezhou Han Population [J]. Current Biotechnology, 2022, 12(2): 288-295. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||