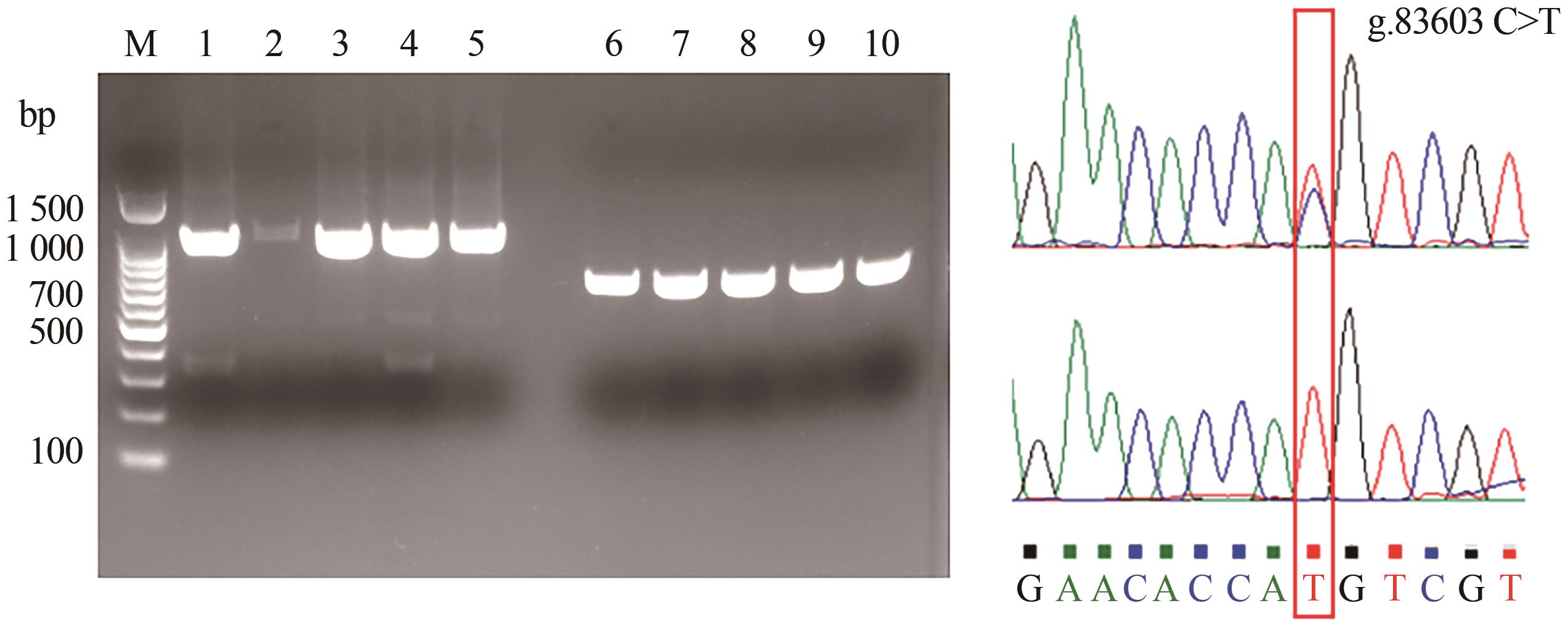
Current Biotechnology ›› 2025, Vol. 15 ›› Issue (1): 119-126.DOI: 10.19586/j.2095-2341.2024.0137
• Articles • Previous Articles Next Articles
Association Analysis Between EPAS1 Gene Polymorphism and Blood Physiological Indicators in Two Different Altitudes of Sheep in Xinjiang
Jiong DONG( ), Shifeng WANG(
), Shifeng WANG( ), Kaidiriye YUSUPU·, Jianli ZHAN, Abudureheman TUERXUN·
), Kaidiriye YUSUPU·, Jianli ZHAN, Abudureheman TUERXUN·
- Key Laboratory of Biological Resources and Ecology of Pamirs Plateau Xinjiang Uygur Autonomous Region,the College of Life and Geographic Sciences,Kashi University,Xinjiang Kashi 844000,China
-
Received:2024-08-21Accepted:2024-10-31Online:2025-01-25Published:2025-03-07 -
Contact:Shifeng WANG
新疆2种不同海拔绵羊EPAS1基因多态性与血液生理指标关联分析
董炅( ), 王世锋(
), 王世锋( ), 凯迪日耶·玉苏普, 詹建立, 阿布都热合曼·吐尔逊
), 凯迪日耶·玉苏普, 詹建立, 阿布都热合曼·吐尔逊
- 喀什大学生命与地理科学学院,新疆帕米尔高原生物资源与生态重点实验室,新疆 喀什 844000
-
通讯作者:王世锋 -
作者简介:董炅E-mail: Dongwith@outlook.com; -
基金资助:新疆高校科研计划项目(XJEDU2021SI024)
CLC Number:
Cite this article
Jiong DONG, Shifeng WANG, Kaidiriye YUSUPU·, Jianli ZHAN, Abudureheman TUERXUN·. Association Analysis Between EPAS1 Gene Polymorphism and Blood Physiological Indicators in Two Different Altitudes of Sheep in Xinjiang[J]. Current Biotechnology, 2025, 15(1): 119-126.
董炅, 王世锋, 凯迪日耶·玉苏普, 詹建立, 阿布都热合曼·吐尔逊. 新疆2种不同海拔绵羊EPAS1基因多态性与血液生理指标关联分析[J]. 生物技术进展, 2025, 15(1): 119-126.
share this article
| 单倍型 | SNP位点(突变频数) | 合计 | ||||||
|---|---|---|---|---|---|---|---|---|
| g.83603(5) | g.83638(48) | g.83734(18) | g.83743(4) | g.91430(3) | g.91692(56) | g.91698(15) | ||
| 合计 | - | - | - | - | - | - | - | 153 |
| NC_056056 | G | T | G | G | C | T | G | 44 |
| Hap1 | A | - | - | - | - | - | - | 2 |
| Hap2 | - | C | - | - | - | - | - | 31 |
| Hap3 | - | - | A | - | - | - | - | 1 |
| Hap4 | - | - | - | A | - | - | - | 1 |
| Hap5 | - | - | - | - | - | C | - | 27 |
| Hap6 | - | - | - | - | - | - | A | 10 |
| Hap7 | A | C | - | - | - | - | - | 1 |
| Hap8 | A | - | G | - | - | - | - | 1 |
| Hap9 | A | - | - | - | - | C | - | 1 |
| Hap10 | - | C | G | - | - | - | - | 1 |
| Hap11 | - | C | - | A | - | - | - | 1 |
| Hap12 | - | C | - | - | - | C | - | 9 |
| Hap13 | - | C | - | - | - | - | A | 3 |
| Hap14 | - | - | G | - | - | C | - | 13 |
| Hap15 | - | - | - | A | - | - | A | 1 |
| Hap16 | - | - | - | - | T | C | - | 3 |
| Hap17 | - | - | - | - | - | C | A | 1 |
| Hap18 | - | - | A | A | - | C | - | 1 |
| Hap19 | - | C | A | - | - | C | - | 1 |
Table 1 SNPs in exons 9 and 16 of the EPAS1 gene and haplotypes
| 单倍型 | SNP位点(突变频数) | 合计 | ||||||
|---|---|---|---|---|---|---|---|---|
| g.83603(5) | g.83638(48) | g.83734(18) | g.83743(4) | g.91430(3) | g.91692(56) | g.91698(15) | ||
| 合计 | - | - | - | - | - | - | - | 153 |
| NC_056056 | G | T | G | G | C | T | G | 44 |
| Hap1 | A | - | - | - | - | - | - | 2 |
| Hap2 | - | C | - | - | - | - | - | 31 |
| Hap3 | - | - | A | - | - | - | - | 1 |
| Hap4 | - | - | - | A | - | - | - | 1 |
| Hap5 | - | - | - | - | - | C | - | 27 |
| Hap6 | - | - | - | - | - | - | A | 10 |
| Hap7 | A | C | - | - | - | - | - | 1 |
| Hap8 | A | - | G | - | - | - | - | 1 |
| Hap9 | A | - | - | - | - | C | - | 1 |
| Hap10 | - | C | G | - | - | - | - | 1 |
| Hap11 | - | C | - | A | - | - | - | 1 |
| Hap12 | - | C | - | - | - | C | - | 9 |
| Hap13 | - | C | - | - | - | - | A | 3 |
| Hap14 | - | - | G | - | - | C | - | 13 |
| Hap15 | - | - | - | A | - | - | A | 1 |
| Hap16 | - | - | - | - | T | C | - | 3 |
| Hap17 | - | - | - | - | - | C | A | 1 |
| Hap18 | - | - | A | A | - | C | - | 1 |
| Hap19 | - | C | A | - | - | C | - | 1 |
| SNP位点 | 基因型 | 基因型频率 | 预期基因型频率 | 等位基因 | 基因频率 | |||
|---|---|---|---|---|---|---|---|---|
| TK(n) | DL(n) | TK | DL | TK | DL | |||
g.83603 G>A 错义突变 | GG | 1.000 0A(79) | 0.932 4B(69) | 1.000 0 | 0.920 6 | G | 1.000 0 | 0.952 5 |
| GA | 0.000 0B(0) | 0.054 1A(4) | 0.000 0 | 0.077 8 | A | 0.000 0 | 0.040 5 | |
| AA | 0.000 0b(0) | 0.013 5a(1) | 0.000 0 | 0.001 6 | ||||
g.83638 T>C 同义突变 | TT | 0.721 5a(57) | 0.648 4b(48) | 0.719 3 | 0.635 7 | T | 0.848 1 | 0.797 3 |
| TC | 0.253 2(20) | 0.297 3(22) | 0.257 7 | 0.323 2 | C | 0.151 9 | 0.202 7 | |
| CC | 0.025 3(2) | 0.054 1(4) | 0.023 1 | 0.041 1 | ||||
g.83734 G>A 同义突变 | GG | 0.873 4(69) | 0.891 9(66) | 0.807 7 | 0.832 0 | G | 0.898 7 | 0.912 2 |
| GA | 0.050 6(4) | 0.040 5(3) | 0.182 0 | 0.160 2 | A | 0.101 3 | 0.087 8 | |
| AA | 0.075 9(6) | 0.067 6(5) | 0.010 3 | 0.007 7 | ||||
g.83743 G>A 同义突变 | GG | 0.949 4B(75) | 1.000 0A(74) | 0.937 7 | 1.000 0 | G | 0.965 4 | 1.000 0 |
| GA | 0.038 0A(3) | 0.000 0B(0) | 0.061 3 | 0.000 0 | A | 0.031 6 | 0.000 0 | |
| AA | 0.012 7a(1) | 0.000 0b(0) | 0.001 0 | 0.000 0 | ||||
g.91430 C>T 同义突变 | TT | 0.000 0(0) | 0.000 0(0) | 0.000 0 | 0.000 2 | T | 0.006 3 | 0.013 5 |
| TC | 0.012 7(1) | 0.027 0(2) | 0.012 6 | 0.026 7 | C | 0.993 7 | 0.986 5 | |
| CC | 0.987 3(78) | 0.973 0(72) | 0.987 4 | 0.973 2 | ||||
g.91692 T>C 同义突变 | TT | 0.632 9(50) | 0.635 1(47) | 0.511 5 | 0.484 3 | T | 0.715 2 | 0.695 9 |
| TC | 0.164 6(13) | 0.121 6(9) | 0.407 4 | 0.423 2 | C | 0.284 8 | 0.304 1 | |
| CC | 0.202 5(16) | 0.243 2(18) | 0.081 1 | 0.092 4 | ||||
g.91698 G>A 同义突变 | GG | 0.911 4(72) | 0.891 9(66) | 0.913 4 | 0.869 4 | G | 0.955 7 | 0.932 4 |
| GA | 0.088 6(7) | 0.081 1(6) | 0.084 7 | 0.126 0 | A | 0.044 3 | 0.067 6 | |
| AA | 0.000 0B(0) | 0.027 0A(2) | 0.002 0 | 0.004 6 | ||||
Table 2 Gene frequency and genotype frequency
| SNP位点 | 基因型 | 基因型频率 | 预期基因型频率 | 等位基因 | 基因频率 | |||
|---|---|---|---|---|---|---|---|---|
| TK(n) | DL(n) | TK | DL | TK | DL | |||
g.83603 G>A 错义突变 | GG | 1.000 0A(79) | 0.932 4B(69) | 1.000 0 | 0.920 6 | G | 1.000 0 | 0.952 5 |
| GA | 0.000 0B(0) | 0.054 1A(4) | 0.000 0 | 0.077 8 | A | 0.000 0 | 0.040 5 | |
| AA | 0.000 0b(0) | 0.013 5a(1) | 0.000 0 | 0.001 6 | ||||
g.83638 T>C 同义突变 | TT | 0.721 5a(57) | 0.648 4b(48) | 0.719 3 | 0.635 7 | T | 0.848 1 | 0.797 3 |
| TC | 0.253 2(20) | 0.297 3(22) | 0.257 7 | 0.323 2 | C | 0.151 9 | 0.202 7 | |
| CC | 0.025 3(2) | 0.054 1(4) | 0.023 1 | 0.041 1 | ||||
g.83734 G>A 同义突变 | GG | 0.873 4(69) | 0.891 9(66) | 0.807 7 | 0.832 0 | G | 0.898 7 | 0.912 2 |
| GA | 0.050 6(4) | 0.040 5(3) | 0.182 0 | 0.160 2 | A | 0.101 3 | 0.087 8 | |
| AA | 0.075 9(6) | 0.067 6(5) | 0.010 3 | 0.007 7 | ||||
g.83743 G>A 同义突变 | GG | 0.949 4B(75) | 1.000 0A(74) | 0.937 7 | 1.000 0 | G | 0.965 4 | 1.000 0 |
| GA | 0.038 0A(3) | 0.000 0B(0) | 0.061 3 | 0.000 0 | A | 0.031 6 | 0.000 0 | |
| AA | 0.012 7a(1) | 0.000 0b(0) | 0.001 0 | 0.000 0 | ||||
g.91430 C>T 同义突变 | TT | 0.000 0(0) | 0.000 0(0) | 0.000 0 | 0.000 2 | T | 0.006 3 | 0.013 5 |
| TC | 0.012 7(1) | 0.027 0(2) | 0.012 6 | 0.026 7 | C | 0.993 7 | 0.986 5 | |
| CC | 0.987 3(78) | 0.973 0(72) | 0.987 4 | 0.973 2 | ||||
g.91692 T>C 同义突变 | TT | 0.632 9(50) | 0.635 1(47) | 0.511 5 | 0.484 3 | T | 0.715 2 | 0.695 9 |
| TC | 0.164 6(13) | 0.121 6(9) | 0.407 4 | 0.423 2 | C | 0.284 8 | 0.304 1 | |
| CC | 0.202 5(16) | 0.243 2(18) | 0.081 1 | 0.092 4 | ||||
g.91698 G>A 同义突变 | GG | 0.911 4(72) | 0.891 9(66) | 0.913 4 | 0.869 4 | G | 0.955 7 | 0.932 4 |
| GA | 0.088 6(7) | 0.081 1(6) | 0.084 7 | 0.126 0 | A | 0.044 3 | 0.067 6 | |
| AA | 0.000 0B(0) | 0.027 0A(2) | 0.002 0 | 0.004 6 | ||||
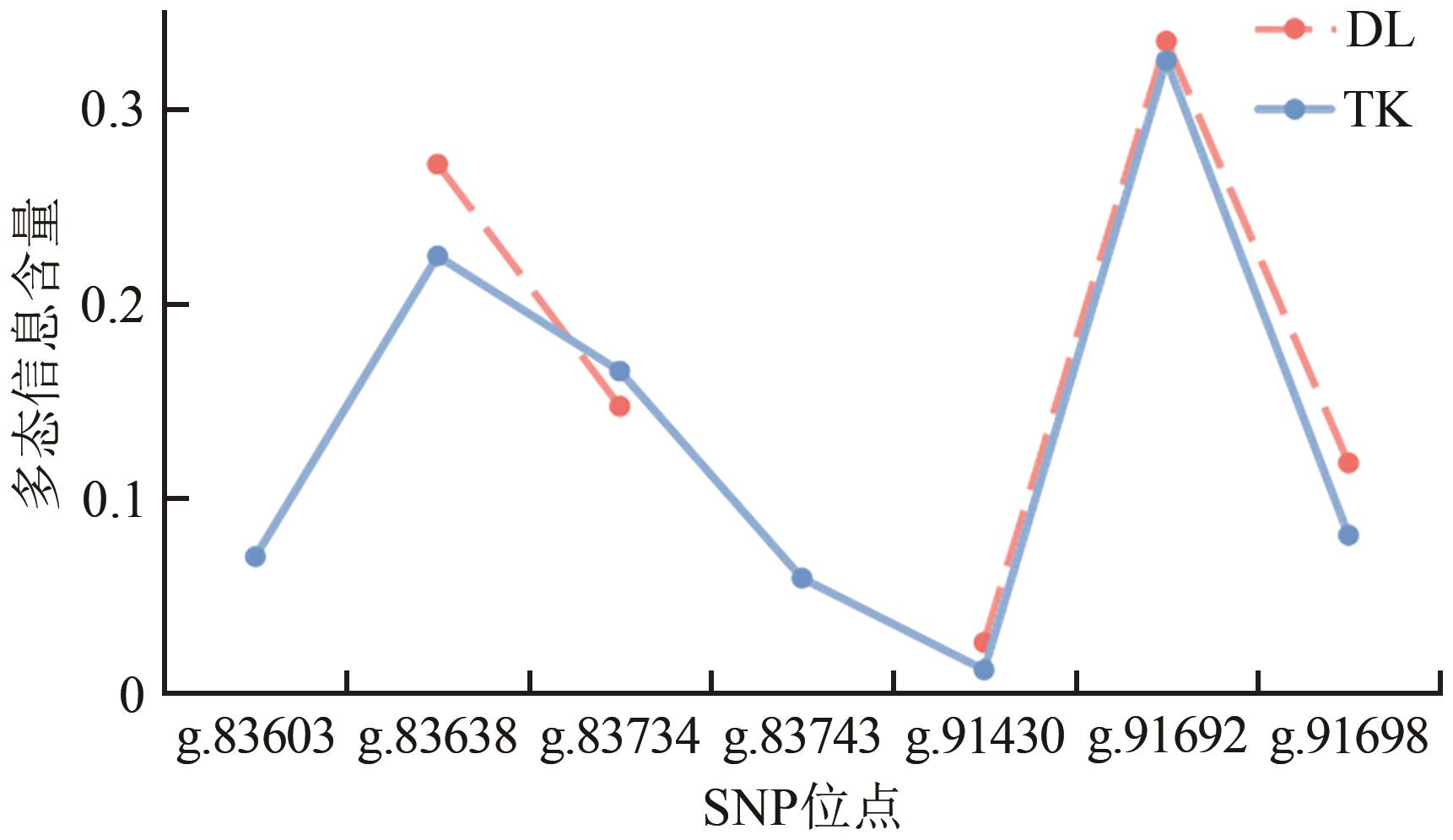
| SNP位点 | 品种 | PIC | He | Ho | Ne | HWE |
|---|---|---|---|---|---|---|
| g.83603 | TK | 0.000 | 0.000 | 1.000 | 1.000 | |
| DL | 0.075 | 0.078 | 0.922 | 1.084 | 0.009 | |
| g.83638 | TK | 0.224 | 0.258 | 0.742 | 1.347 | 0.877 |
| DL | 0.271 | 0.323 | 0.677 | 1.478 | 0.490 | |
| g.83734 | TK | 0.165 | 0.182 | 0.818 | 1.223 | 0.000 |
| DL | 0.147 | 0.160 | 0.840 | 1.191 | 0.000 | |
| g.83743 | TK | 0.059 | 0.061 | 0.939 | 1.065 | 0.001 |
| DL | 0.000 | 0.000 | 1.000 | 1.000 | ||
| g.91430 | TK | 0.012 | 0.013 | 0.987 | 1.013 | 0.955 |
| DL | 0.026 | 0.027 | 0.973 | 1.027 | 0.906 | |
| g.91692 | TK | 0.324 | 0.407 | 0.593 | 1.687 | 0.000 |
| DL | 0.334 | 0.423 | 0.577 | 1.734 | 0.000 | |
| g.91698 | TK | 0.081 | 0.085 | 0.915 | 1.093 | 0.680 |
| DL | 0.118 | 0.126 | 0.874 | 1.144 | 0.002 |
Table 3 Population genetics parameters and the Hardy-Weinberg law test
| SNP位点 | 品种 | PIC | He | Ho | Ne | HWE |
|---|---|---|---|---|---|---|
| g.83603 | TK | 0.000 | 0.000 | 1.000 | 1.000 | |
| DL | 0.075 | 0.078 | 0.922 | 1.084 | 0.009 | |
| g.83638 | TK | 0.224 | 0.258 | 0.742 | 1.347 | 0.877 |
| DL | 0.271 | 0.323 | 0.677 | 1.478 | 0.490 | |
| g.83734 | TK | 0.165 | 0.182 | 0.818 | 1.223 | 0.000 |
| DL | 0.147 | 0.160 | 0.840 | 1.191 | 0.000 | |
| g.83743 | TK | 0.059 | 0.061 | 0.939 | 1.065 | 0.001 |
| DL | 0.000 | 0.000 | 1.000 | 1.000 | ||
| g.91430 | TK | 0.012 | 0.013 | 0.987 | 1.013 | 0.955 |
| DL | 0.026 | 0.027 | 0.973 | 1.027 | 0.906 | |
| g.91692 | TK | 0.324 | 0.407 | 0.593 | 1.687 | 0.000 |
| DL | 0.334 | 0.423 | 0.577 | 1.734 | 0.000 | |
| g.91698 | TK | 0.081 | 0.085 | 0.915 | 1.093 | 0.680 |
| DL | 0.118 | 0.126 | 0.874 | 1.144 | 0.002 |
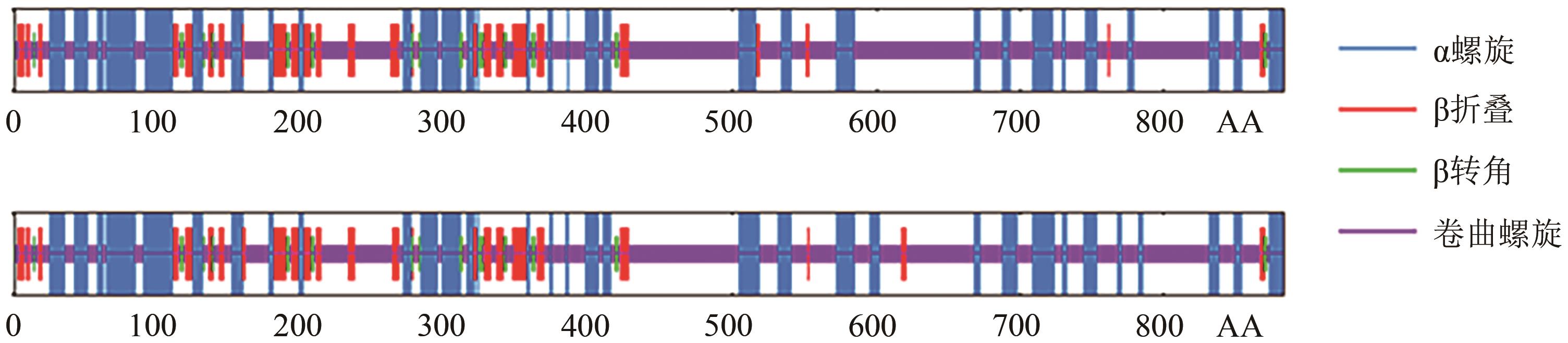
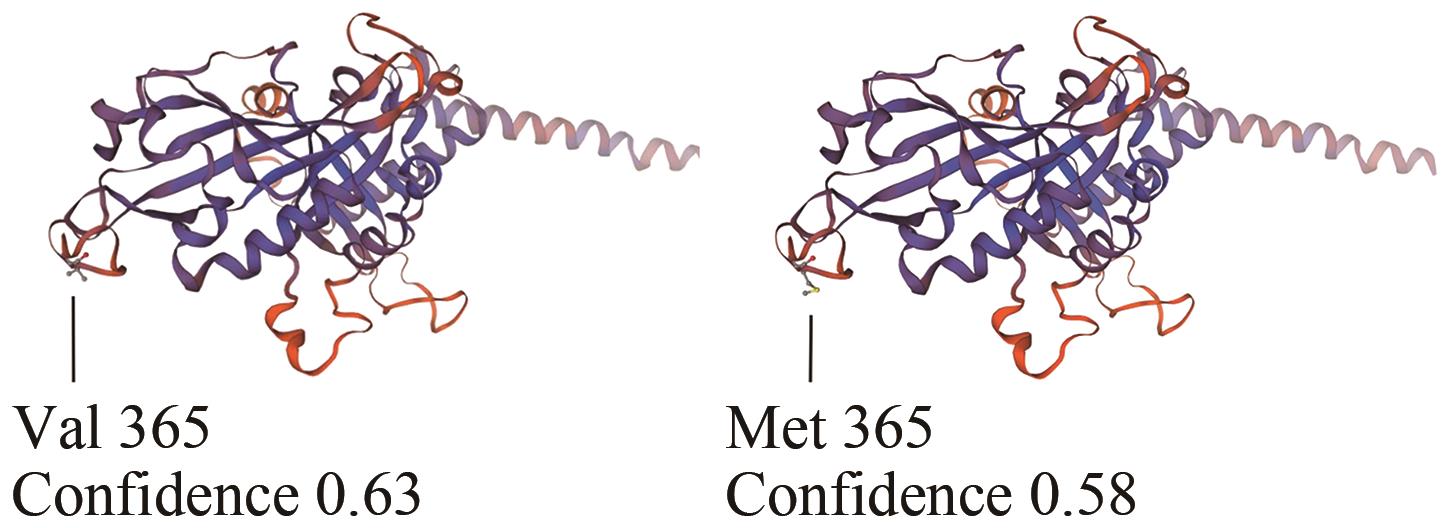
| 种类 | 突变前 | 突变后 |
|---|---|---|
| 缬氨酸占比 | 4.1% | 4.0% |
| 甲硫氨酸占比 | 3.4% | 3.5% |
| 化学式 | C4283H6688N1162O1341S52 | C4283H6688N1162O1341S53 |
| 原子总数 | 13 526 | 13 527 |
| 消光系数(胱氨酸)1 | 0.719 | 0.718 |
| 消光系数(半胱氨酸)2 | 0.705 | 0.704 |
| 脂溶指数 | 67.61 | 67.28 |
| 亲水平均值 | -0.479 | -0.482 |
Table 4 Physicochemical properties of proteins before and after mutation
| 种类 | 突变前 | 突变后 |
|---|---|---|
| 缬氨酸占比 | 4.1% | 4.0% |
| 甲硫氨酸占比 | 3.4% | 3.5% |
| 化学式 | C4283H6688N1162O1341S52 | C4283H6688N1162O1341S53 |
| 原子总数 | 13 526 | 13 527 |
| 消光系数(胱氨酸)1 | 0.719 | 0.718 |
| 消光系数(半胱氨酸)2 | 0.705 | 0.704 |
| 脂溶指数 | 67.61 | 67.28 |
| 亲水平均值 | -0.479 | -0.482 |
| 项目 | GG | GA | AA |
|---|---|---|---|
| 白细胞数/(109·L-1) | 7.83±2.04 | 8.42±4.36 | 5.56 |
| 红细胞数/(109·L-1) | 8.19±1.02 | 8.31±0.86 | 6.04 |
| 血小板数/(109·L-1) | 304.38±155.43 | 215.00±118.79 | 331.00 |
| 淋巴细胞数/(109·L-1) | 2.29±1.25 | 2.54±0.23 | 0.83 |
| 单核细胞数/(109·L-1) | 0.33±0.15 | 0.48±0.02 | 0.37 |
| 中性粒细胞数/(109·L-1) | 5.10±1.84 | 5.28±4.09 | 4.25 |
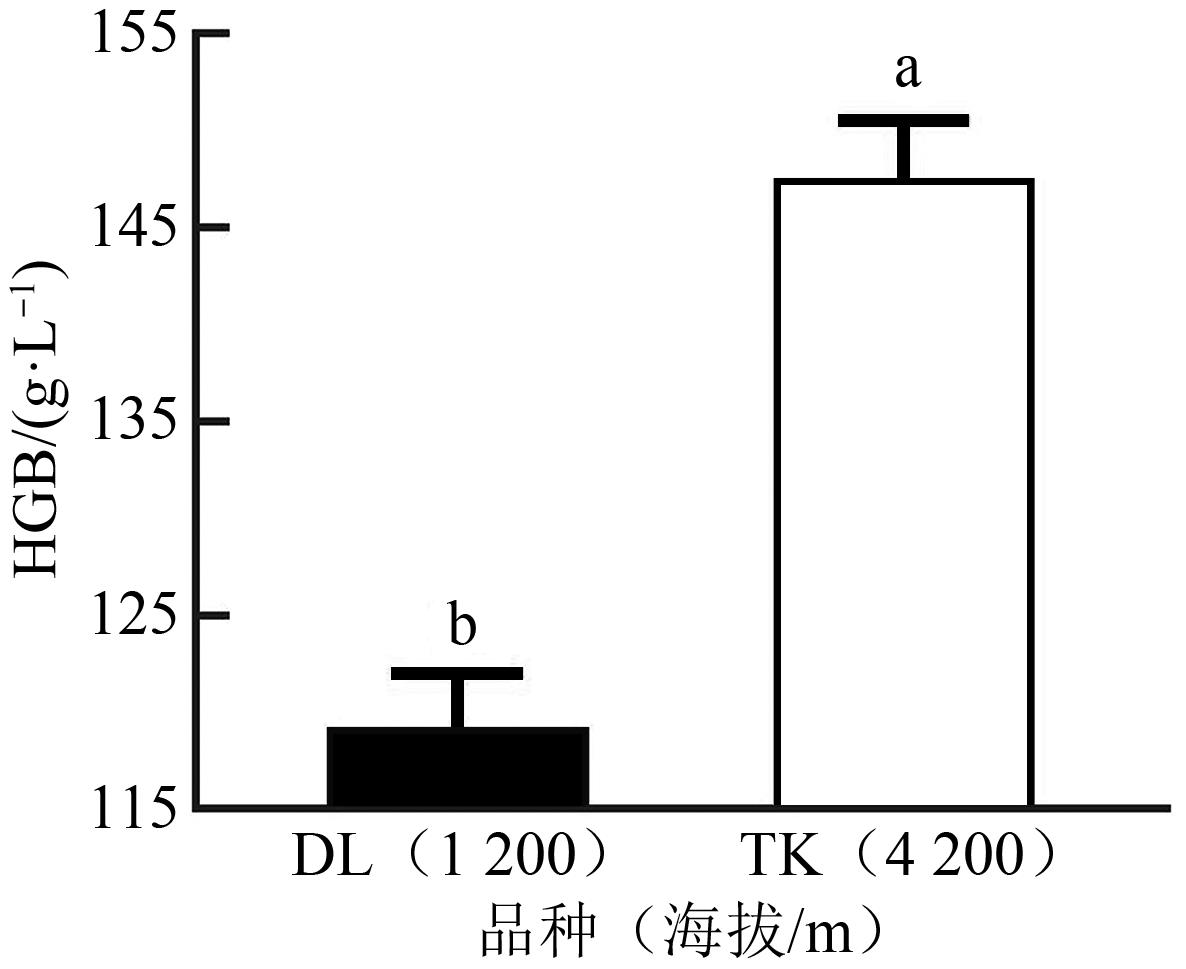
| 血红蛋白含量/(g·L-1) | 101.38±16.46 | 101.5±13.44 | 74.00 |
| 平均红细胞血红蛋白浓度/(g·L-1) | 355.50±25.22 | 370.50±12.02 | 316.00 |
| 红细胞压积/% | 28.67±4.47 | 27.4±2.69 | 23.50 |
| 嗜酸性粒细胞百分比/% | 1.48±1.09 | 1.95±1.06 | 2.00 |
| 红细胞分布宽度变异系数/% | 16.78±1.08 | 16.95±2.05 | 16.10 |
| 血小板分布宽度/fL | 8.43±1.65 | 9.15±0.78 | 4.80 |
| 平均血小板体积/fL | 5.17±0.61 | 5.45±0.49 | 4.80 |
| 平均红细胞体积/fL | 34.88±2.16 | 33.00±0.14 | 38.90 |
| 红细胞分布宽度标准差/fL | 21.45±2.29 | 20.15±1.77 | 23.70 |
Table 5 The blood physiological indexes in different genotypes
| 项目 | GG | GA | AA |
|---|---|---|---|
| 白细胞数/(109·L-1) | 7.83±2.04 | 8.42±4.36 | 5.56 |
| 红细胞数/(109·L-1) | 8.19±1.02 | 8.31±0.86 | 6.04 |
| 血小板数/(109·L-1) | 304.38±155.43 | 215.00±118.79 | 331.00 |
| 淋巴细胞数/(109·L-1) | 2.29±1.25 | 2.54±0.23 | 0.83 |
| 单核细胞数/(109·L-1) | 0.33±0.15 | 0.48±0.02 | 0.37 |
| 中性粒细胞数/(109·L-1) | 5.10±1.84 | 5.28±4.09 | 4.25 |
| 血红蛋白含量/(g·L-1) | 101.38±16.46 | 101.5±13.44 | 74.00 |
| 平均红细胞血红蛋白浓度/(g·L-1) | 355.50±25.22 | 370.50±12.02 | 316.00 |
| 红细胞压积/% | 28.67±4.47 | 27.4±2.69 | 23.50 |
| 嗜酸性粒细胞百分比/% | 1.48±1.09 | 1.95±1.06 | 2.00 |
| 红细胞分布宽度变异系数/% | 16.78±1.08 | 16.95±2.05 | 16.10 |
| 血小板分布宽度/fL | 8.43±1.65 | 9.15±0.78 | 4.80 |
| 平均血小板体积/fL | 5.17±0.61 | 5.45±0.49 | 4.80 |
| 平均红细胞体积/fL | 34.88±2.16 | 33.00±0.14 | 38.90 |
| 红细胞分布宽度标准差/fL | 21.45±2.29 | 20.15±1.77 | 23.70 |
| 1 | GORDAN J D, BERTOUT J A, HU C J, et al.. HIF-2α promotes hypoxic cell proliferation by enhancing c-Myc transcriptional activity[J]. Cancer Cell, 2007, 11(4): 335-347. |
| 2 | HOLMQUIST-MENGELBIER L, FREDLUND E, LÖFSTEDT T, et al.. Recruitment of HIF-1alpha and HIF-2alpha to common target genes is differentially regulated in neuroblastoma:HIF-2alpha promotes an aggressive phenotype[J]. Cancer cell,2006, 10(5): 413-423. |
| 3 | 高文宇, 曾蓉, 许梦娜, 等. EPAS1基因rs1868092位点多态性与中国藏族人群高原红细胞增多症的相关性研究[J]. 大理大学学报, 2023, 8(4): 39-42. |
| GAO W Y, ZENG R, XU M N, et al.. Study on the correlation between polymorphism of EPAS1 gene rs1868092 and high altitude polycythemia in Tibetan population[J]. J. Dali Univ., 2023, 8(4): 39-42. | |
| 4 | 张烨, 蒋荣珍. HIF2α与细胞通透性关系的研究进展[J].医学研究杂志, 2018, 47(1): 13-15+114. |
| 5 | SKULI N, LIU L, RUNGE A, et al.. Endothelial deletion of hypoxia-inducible factor-2α (HIF-2α) alters vascular function and tumor angiogenesis[J]. Blood, 2009, 114(2): 469-477. |
| 6 | SOWTER H M, RAVAL R, MOORE J, et al.. Predominant role of hypoxia-inducible transcription factor (Hif)-1α versus Hif-2α in regulation of the transcriptional response to hypoxia[J]. Cancer Res., 2003, 63(19): 6130-6134. |
| 7 | 张欣, 韩旭. 缺氧诱导因子2在妇科恶性肿瘤中表达的研究进展[J]. 中国医药, 2021, 16(10): 1593-1596. |
| ZHANG X, HAN X. Research progress of hypoxia-inducible factor-2 expression in gynecological malignancies[J]. China Med., 2021, 16(10): 1593-1596. | |
| 8 | FRIEDRICH J, WIENER P. Selection signatures for high-altitude adaptation in ruminants[J]. Anim. Genet., 2020, 51(2): 157-165. |
| 9 | 万星, 刘雅婧, 黄家辉, 等. 反刍家畜高海拔低氧适应性性状的鉴定研究现状与展望[J].中国科学: 生命科学, 2023, 53(7): 964-980. |
| WANG X, LIU Y J, HUANG J H, et al.. Identification methods for high-altitude adaptation of phenotypes in ruminant livestock: status and prospect[J]. Sci. Sin., 2023, 53(7): 964-980. | |
| 10 | ZHANG Y Y, XUE X L, LIU Y, et al.. Genome-wide comparative analyses reveal selection signatures underlying adaptation and production in Tibetan and Poll Dorset sheep[J]. Sci. Rep., 2021, 11: 2466[2025-02-01]. . |
| 11 | 索南元旦. 关于藏绵羊高海拔地区适应性及生产性能研究[J].中国动物保健, 2022, 24(8): 77-78. |
| 12 | 孙晓萍, 刘建斌, 袁超, 等. 藏绵羊高海拔适应性及生产性能分析[J]. 今日畜牧兽医, 2018, 34(4): 50-52. |
| 13 | 宋伸. 多组学研究揭示山羊高海拔适应性及产绒性状分子机制[D]. 北京: 中国农业大学, 2017. |
| 14 | 朱莉, 李根, 孔小艳, 等. 藏绵羊血红蛋白、EPAS1基因与低氧适应相关性研究[J]. 云南农业大学学报(自然科学), 2020, 35(3): 436-442+518. |
| ZHU L, LI G, KONG X Y, et al.. The association of genes hemoglobin and EPAS1 with hypoxia adaptation in the Tibetan Sheep[J]. J. Yunnan Agric. Univ. (Nat. Sci.), 2020, 35(3):436-442+518. | |
| 15 | 包鹏甲, 裴杰, 阎萍, 等. 藏羊EPO基因遗传多样性与高原低氧适应性[J]. 江苏农业学报, 2014, 30(3): 581-585. |
| BAO P J, PEI J, YAN P, et al.. EPO gene polymorphism and high altitude hypoxia adaptation of Tibetan sheep[J]. Jiangsu J.Agric. Sci., 2014, 30(3): 581-585. | |
| 16 | 白晶晶, 刘秀. 藏绵羊低氧适应的血液生理指标特征研究[J].青海畜牧兽医杂志, 2023, 53(1): 19-22. |
| BAI J J, LIU X. Study on characteristics of blood physiological indexes of hypoxia adaptation in Tibetan sheep[J]. Chin. Qinghai J. Anim. Vet. Sci., 2023, 53(1): 19-22. | |
| 17 | 雷艳, 蔡鹏, 兰斌, 等. 饲养在平原与高原的塔什库尔干羊血液生理生化指标比较[J]. 新疆畜牧业, 2023, 39(1):16-18+47. |
| 18 | 卢晓丽, 赵彦玲, 吴征王, 等.西藏色瓦藏绵羊高原适应性的血液生理学特性研究[J].西南农业学报, 2019, 32(6): 1443-1447. |
| LU X L, ZHAO Y L, WU Z W, et al.. Study on blood physiological characteristics of plateau adaptation in Tibetan sawa sheep[J]. Southwest China J. Agric. Sci., 2019, 32(6): 1443-1447. | |
| 19 | 李孝仪, 杨舒黎, 马黎, 等. 西藏绵羊低氧适应的血液生理学特性研究[J]. 黑龙江畜牧兽医(上半月), 2016(1): 12-15. |
| LI X Y, YANG S L, MA L, et al.. Study on the blood physiological properties of hypoxia adaptation in Tibetan sheep[J]. Heilongjiang Anim. Sci. Vet. Med., 2016(1): 12-15. | |
| 20 | 张春梅, 陈倩, 谢雯, 等. 基于促红细胞生成素基因表达及血液生理生化指标探究湖羊在青海高海拔地区的适应性[J]. 动物营养学报, 2020, 32(11): 5380-5387. |
| ZHANG C M, CHEN Q, XIE W, et al.. Based on erythropoietin gene expression and blood physiological and biochemical indexes to explore the adaptability of hu sheep in high-altitude area of Qinghai[J]. Chin. J. Anim. Nutr., 2020, 32(11): 5380-5387. | |
| 21 | 高乐程, 巴桑珠扎, 班旦, 等. 西藏改良奶牛低氧适应的血液生理指标与生产性能分析[J]. 饲料研究, 2024, 47(8): 92-96. |
| GAO L C, BASANG Z Z, BAN D, et al.. Analysis on blood physiological indexes and production performance of hypoxic adaptation of Xizang improved dairy cows[J]. Feed Res., 2024, 47(8): 92-96. | |
| 22 | 赵雪, 魏雪锋, 连林生, 等. 藏马低氧适应的血液生理指标研究[J]. 云南农业大学学报(自然科学), 2014, 29(5): 684-688. |
| ZHAO X, WEI X F, LIAN L S, et al.. Study on blood physiological indicators of adaptation to hypoxia in Tibet horse[J]. J. Yunnan Agric. Univ. (Nat. Sci.), 2014, 29(5): 684-688. | |
| 23 | 黄文超. 高原红细胞增多与高血粘度发生的相关分析[J]. 西南军医, 2010, 12(6): 1047-1048. |
| HUANG W C. Correlation analysis of polycythemia and the occurrence of high blood viscosity at high altitude[J]. J. Mil. Surg. Southwest China, 2010, 12(6): 1047-1048. |
| [1] | Luyao LI, Ning LYU, Yongjie ZHAI, Run ZHANG, Anxin YAN, Fuquan JIA. Genetic Polymorphism Analysis on Short Tandem Repeats of Y Chromosome in Han Population of Hohhot [J]. Current Biotechnology, 2025, 15(3): 544-554. |
| [2] | Wenhao ZHAO, He ZHAI. Research Progress on the Relationship Between Genes and Main Behavioral Traits in Working Dogs [J]. Current Biotechnology, 2024, 14(5): 761-767. |
| [3] | Longfei MIAO, Dapeng SUN, Wenhua MA, Jianjun ZHONG. Analysis of Genetic Polymorphisms at 37 Y⁃STR Loci in Dezhou Han Population [J]. Current Biotechnology, 2022, 12(2): 288-295. |
| [4] | LI Sheng-yan, LANG Zhi-hong*, HUANG Da-fang*. Research Progress on Eukaryotic Promoter [J]. Curr. Biotech., 2014, 4(3): 158-164. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||