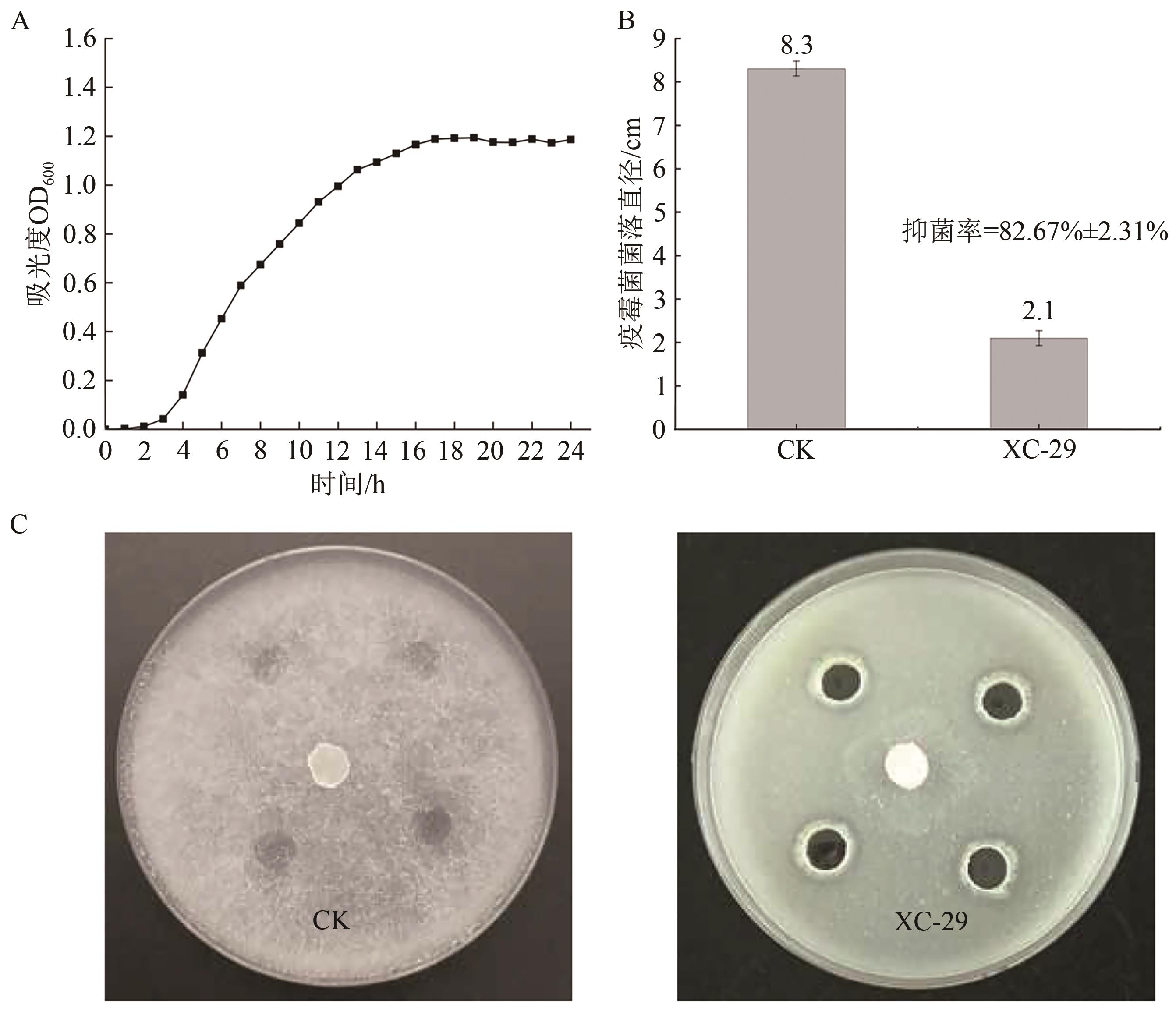
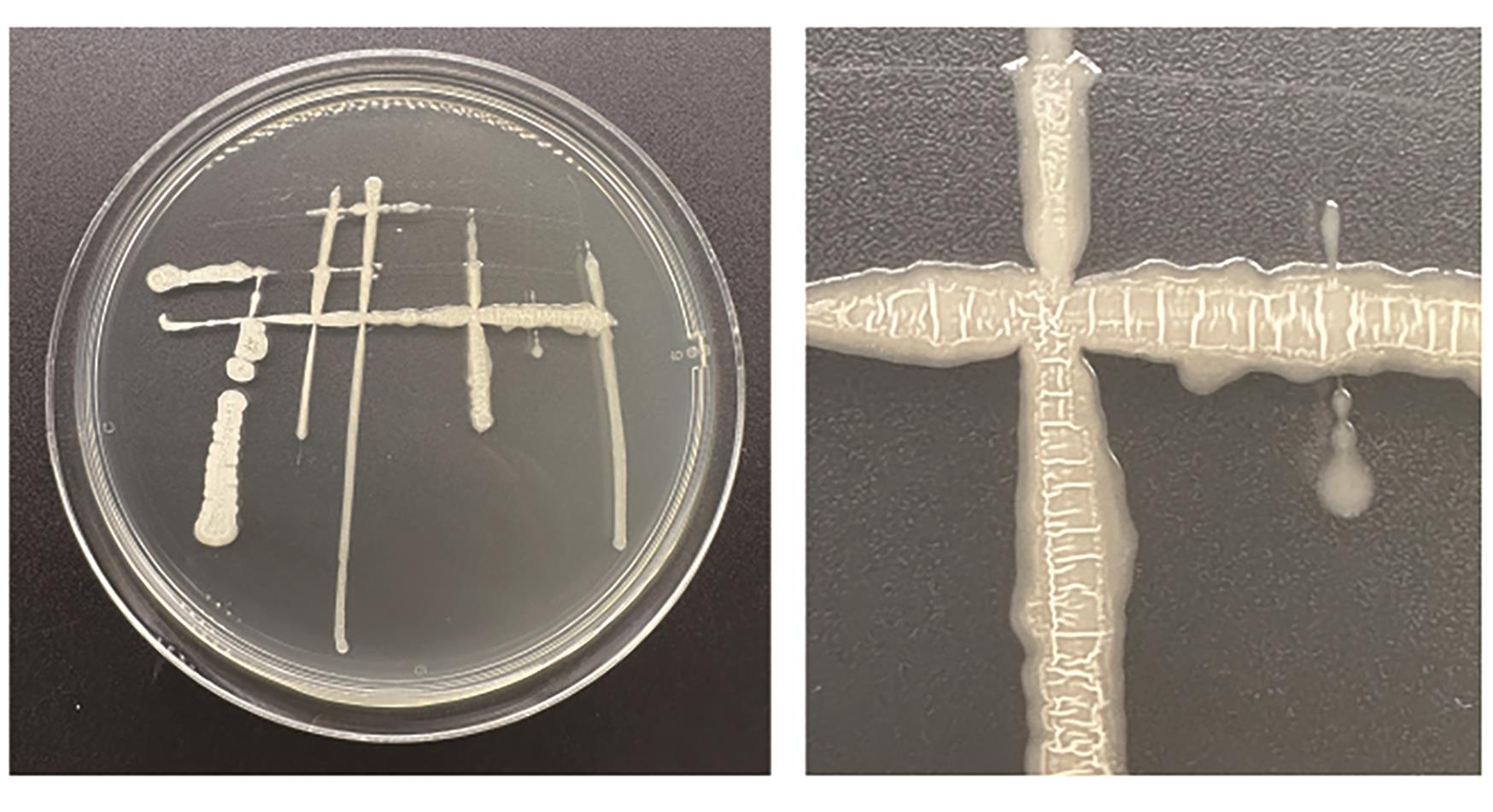
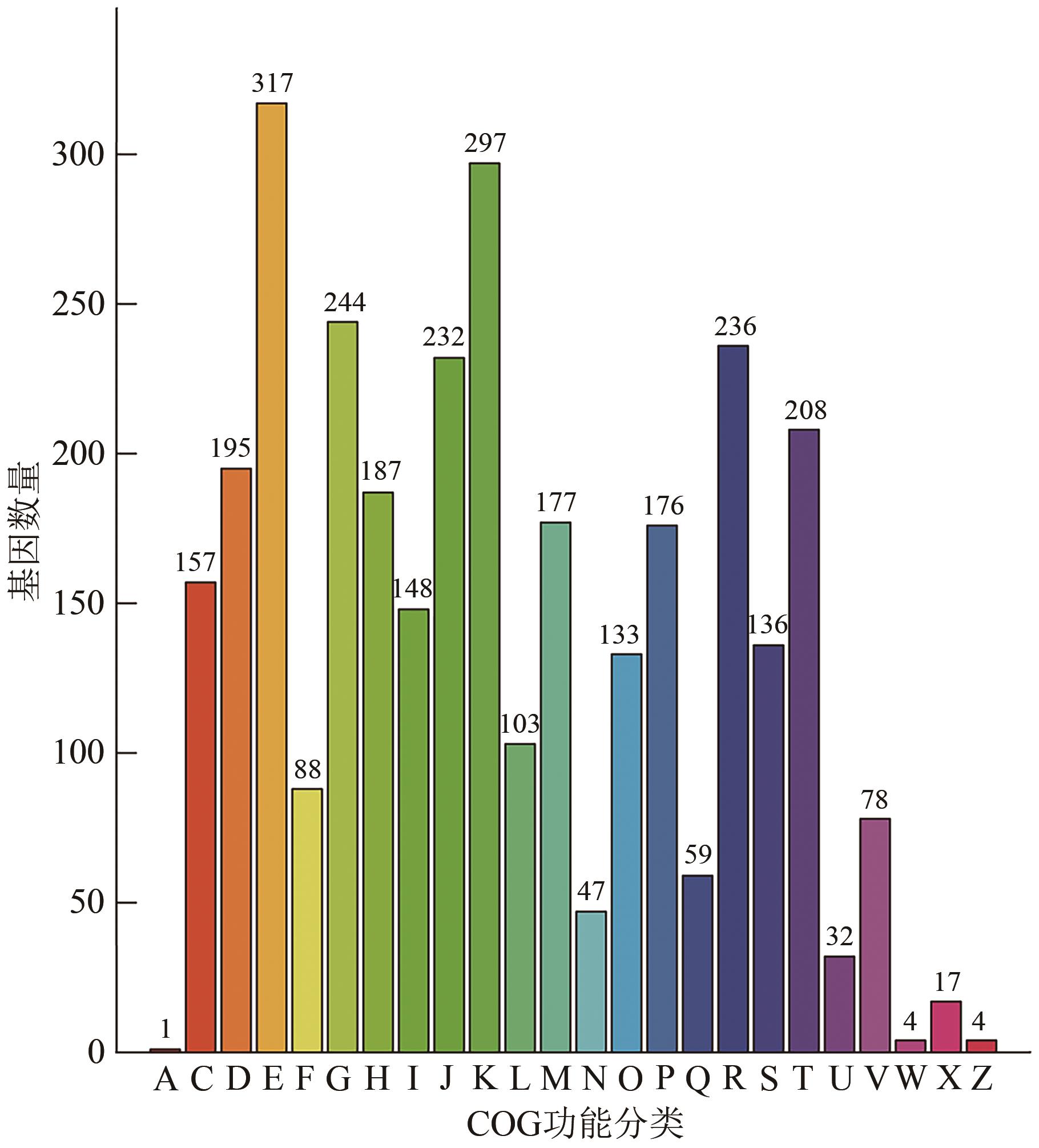
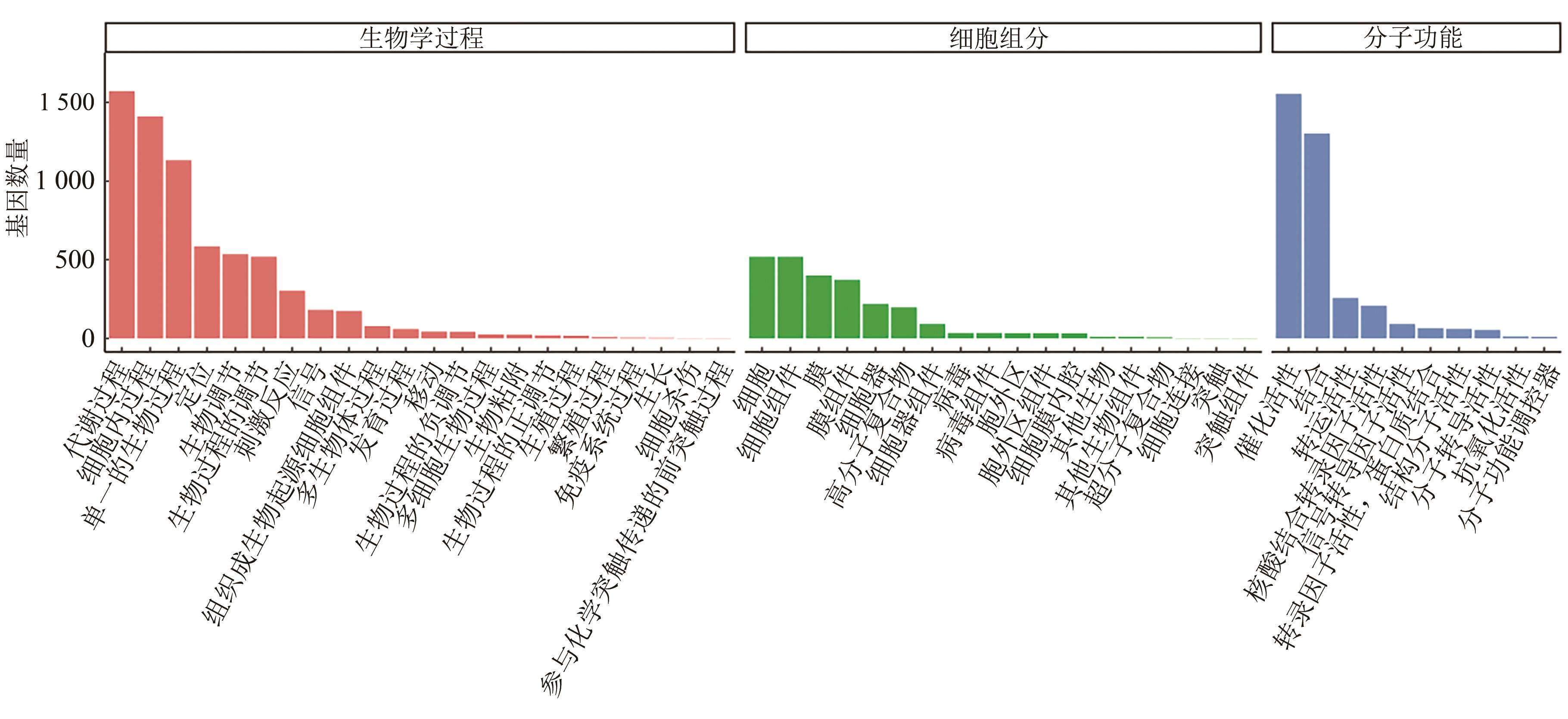
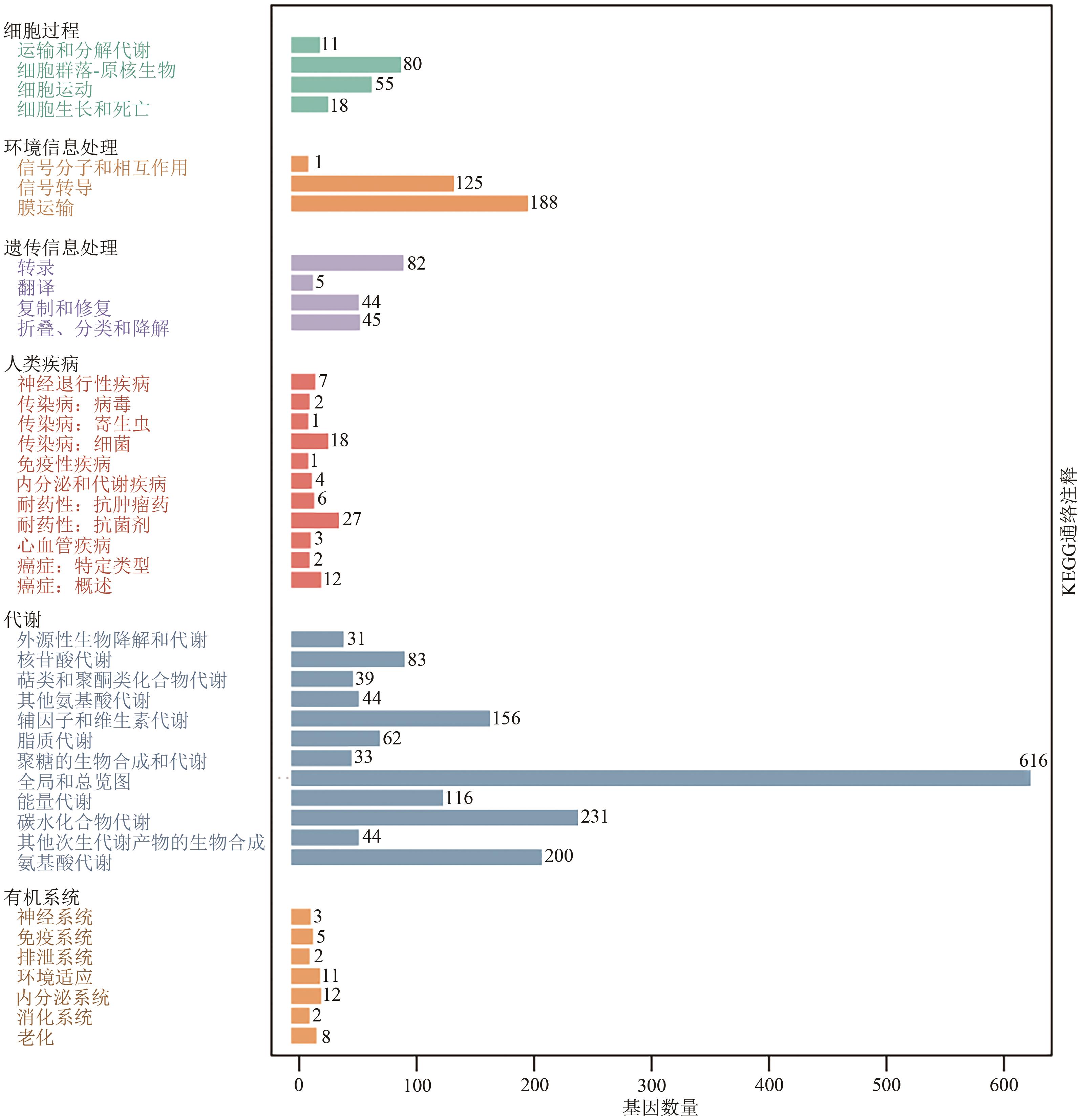
Current Biotechnology ›› 2024, Vol. 14 ›› Issue (6): 1004-1015.DOI: 10.19586/j.2095-2341.2024.0086
• Articles • Previous Articles Next Articles
Isolation, Identification and Whole-genome Sequence Analysis of Phytophthora nicotianae Antagonistic Bacteria XC-29
Sujie WANG1( ), Mengli GU1, Jiemeng TAO1,2, Zhijun TONG3, Junjia GUO1, Jingjing JIN1,2, Mengxiao XU1, Lijun MENG1, Jianfeng ZHANG1,2, Peijian CAO1,2, Peng LU1,2(
), Mengli GU1, Jiemeng TAO1,2, Zhijun TONG3, Junjia GUO1, Jingjing JIN1,2, Mengxiao XU1, Lijun MENG1, Jianfeng ZHANG1,2, Peijian CAO1,2, Peng LU1,2( )
)
- 1.Zhengzhou Tobacco Research Institute of China National Tobacco Corporation,Zhengzhou 450001,China
2.Beijing Life Science Academy,Beijing 102200,China
3.Yunan Academy of Tobacco Agricultural Sciences,Kunming 650106,China
-
Received:2024-04-17Accepted:2024-06-21Online:2024-11-25Published:2024-12-27 -
Contact:Peng LU
烟草疫霉拮抗菌XC-29的分离鉴定及其全基因组序列分析
汪苏洁1( ), 顾梦丽1, 陶界锰1,2, 童治军3, 郭俊佳1, 金静静1,2, 徐梦晓1, 孟利军1, 张剑锋1,2, 曹培健1,2, 卢鹏1,2(
), 顾梦丽1, 陶界锰1,2, 童治军3, 郭俊佳1, 金静静1,2, 徐梦晓1, 孟利军1, 张剑锋1,2, 曹培健1,2, 卢鹏1,2( )
)
- 1.中国烟草总公司郑州烟草研究院,郑州 450001
2.北京生命科技研究院,北京 102200
3.云南省烟草农业科学研究院,昆明 650106
-
通讯作者:卢鹏 -
作者简介:汪苏洁 E-mail: wangsujie951230@163.com; -
基金资助:河南省科技攻关项目(212102110257);中国烟草总公司重点研发项目〔110202102050;110202201001(JY-01)〕;郑州烟草研究院省席科学家项目(902023CK0890)
CLC Number:
Cite this article
Sujie WANG, Mengli GU, Jiemeng TAO, Zhijun TONG, Junjia GUO, Jingjing JIN, Mengxiao XU, Lijun MENG, Jianfeng ZHANG, Peijian CAO, Peng LU. Isolation, Identification and Whole-genome Sequence Analysis of Phytophthora nicotianae Antagonistic Bacteria XC-29[J]. Current Biotechnology, 2024, 14(6): 1004-1015.
汪苏洁, 顾梦丽, 陶界锰, 童治军, 郭俊佳, 金静静, 徐梦晓, 孟利军, 张剑锋, 曹培健, 卢鹏. 烟草疫霉拮抗菌XC-29的分离鉴定及其全基因组序列分析[J]. 生物技术进展, 2024, 14(6): 1004-1015.
share this article
| 类型 | 特征 | 数值 |
|---|---|---|
| 基因组 | 染色体个数 | 1 |
| 基因组序列总长度/bp | 3 719 058 | |
| 编码蛋白基因 | 基因数 | 3 870 |
| 基因总长度/bp | 3 310 209 | |
| 基因平均长度/bp | 855 | |
| 基因序列中GC含量/% | 42.24 | |
| 基因间区 | 基因间区总长度/bp | 408 849 |
| 基因间区GC含量/% | 37.87 | |
| 非编码 | tRNA拷贝数 | 81 |
| 5S rRNA拷贝数 | 8 | |
| 16S rRNA拷贝数 | 8 | |
| 23S rRNA拷贝数 | 8 | |
| sRNA拷贝数 | 7 |
Table 1 Whole genome sequence analysis of biocontrol strain XC-29
| 类型 | 特征 | 数值 |
|---|---|---|
| 基因组 | 染色体个数 | 1 |
| 基因组序列总长度/bp | 3 719 058 | |
| 编码蛋白基因 | 基因数 | 3 870 |
| 基因总长度/bp | 3 310 209 | |
| 基因平均长度/bp | 855 | |
| 基因序列中GC含量/% | 42.24 | |
| 基因间区 | 基因间区总长度/bp | 408 849 |
| 基因间区GC含量/% | 37.87 | |
| 非编码 | tRNA拷贝数 | 81 |
| 5S rRNA拷贝数 | 8 | |
| 16S rRNA拷贝数 | 8 | |
| 23S rRNA拷贝数 | 8 | |
| sRNA拷贝数 | 7 |
| 类型 | 基因数 | 占总基因百分比 |
|---|---|---|
| COG | 2 896 | 74.83% |
| GO | 2 708 | 69.97% |
| KEGG | 3 652 | 94.37% |
| CAZy | 136 | 3.51% |
Table 2 Database distribution of gene functional annotation from the biocontrol bacterium XC-29
| 类型 | 基因数 | 占总基因百分比 |
|---|---|---|
| COG | 2 896 | 74.83% |
| GO | 2 708 | 69.97% |
| KEGG | 3 652 | 94.37% |
| CAZy | 136 | 3.51% |
| 通路编号 | 类目 | 基因数(>40) |
|---|---|---|
| Map01100 | 代谢途径 | 566 |
| Map01110 | 次生代谢物的生物合成 | 276 |
| Map01130 | 抗生素的生物合成 | 211 |
| Map01120 | 不同环境下的微生物代谢 | 170 |
| Map02010 | ABC转运蛋白 | 138 |
| Map01230 | 氨基酸的生物合成 | 130 |
| Map02020 | 双组分调节系统 | 111 |
| Map01200 | 碳代谢 | 98 |
| Map02024 | 群体效应 | 70 |
| Map00230 | 嘌呤代谢 | 59 |
| Map03010 | 核糖体 | 52 |
| Map00240 | 嘧啶代谢 | 45 |
| Map00010 | 糖酵解/糖异生 | 44 |
| Map00620 | 丙酮酸代谢 | 44 |
| Map00270 | 半胱氨酸与蛋氨酸代谢 | 40 |
Table 3 Main metabolic pathways of the biocontrol bacterium XC-29 from KEGG
| 通路编号 | 类目 | 基因数(>40) |
|---|---|---|
| Map01100 | 代谢途径 | 566 |
| Map01110 | 次生代谢物的生物合成 | 276 |
| Map01130 | 抗生素的生物合成 | 211 |
| Map01120 | 不同环境下的微生物代谢 | 170 |
| Map02010 | ABC转运蛋白 | 138 |
| Map01230 | 氨基酸的生物合成 | 130 |
| Map02020 | 双组分调节系统 | 111 |
| Map01200 | 碳代谢 | 98 |
| Map02024 | 群体效应 | 70 |
| Map00230 | 嘌呤代谢 | 59 |
| Map03010 | 核糖体 | 52 |
| Map00240 | 嘧啶代谢 | 45 |
| Map00010 | 糖酵解/糖异生 | 44 |
| Map00620 | 丙酮酸代谢 | 44 |
| Map00270 | 半胱氨酸与蛋氨酸代谢 | 40 |
| 类型 | 家族成员 | 基因数 |
|---|---|---|
| AA | AA1 | 1 |
| CBM | CBM12、CBM13、CBM48、CBM50 | 35 |
| CE | CE0、CE12、CE14、CE4、CE7、CE8、CE9 | 18 |
| GH | GH0、GH1、GH3、GH4、GH5、GH9、GH10、GH11、GH13、GH16、GH18、GH23、GH28、GH30、GH32、GH33、GH38、GH42、GH43、GH48、GH51、GH53、GH73、GH101、GH105、GH126、GH170、GH171 | 55 |
| GT | GT0、GT1、GT4、GT9、GT26、GT28、GT30、GT51、GT58 | 28 |
| PL | PL1、PL9 | 2 |
Table 4 CAZy database function annotation of biocontrol bacterium XC-29 genome
| 类型 | 家族成员 | 基因数 |
|---|---|---|
| AA | AA1 | 1 |
| CBM | CBM12、CBM13、CBM48、CBM50 | 35 |
| CE | CE0、CE12、CE14、CE4、CE7、CE8、CE9 | 18 |
| GH | GH0、GH1、GH3、GH4、GH5、GH9、GH10、GH11、GH13、GH16、GH18、GH23、GH28、GH30、GH32、GH33、GH38、GH42、GH43、GH48、GH51、GH53、GH73、GH101、GH105、GH126、GH170、GH171 | 55 |
| GT | GT0、GT1、GT4、GT9、GT26、GT28、GT30、GT51、GT58 | 28 |
| PL | PL1、PL9 | 2 |
| 基因簇 | 次级代谢产物类型 | 基因数量 | 相似基因簇 | 相似度 |
|---|---|---|---|---|
| 簇1 | 非核糖体肽合成酶 | 43 | 地衣素 | BGC0000381(92%) |
| 簇2 | 非核糖体肽合成酶、Ⅰ型聚酮类化合物 | 48 | 双效菌素 | BGC0001059(18%) |
| 簇3 | 包含Rev响应元件 | 24 | 铁载体二异羟肟酸酯 | - |
| 簇4 | 萜类化合物、镍-铁载体 | 27 | - | BGC0002683(60%) |
| 簇5 | β-内酯 | 24 | 丰原素 | BGC0001095(53%) |
| 簇6 | 萜类化合物、镍-铁载体 | 21 | - | - |
| 簇7 | Ⅲ型聚酮类化合物 | 47 | - | - |
| 簇8 | 核糖体合成和翻译后修饰肽类似物 | 15 | - | - |
| 簇9 | β-内酯 | 33 | - | - |
| 簇10 | 其他 | 46 | 杆菌溶素 | BGC0001184(85%) |
| 簇11 | 非核糖体肽-载体、非核糖体肽合成酶 | 39 | 嗜铁素bacilibactin(E/F) | BGC0002695(80%) |
| 嗜铁素bacilibactin | BGC0000309(100%) | |||
| 嗜铁素paenibactin | BGC0000401(100%) | |||
| 嗜铁素bacilibactin | BGC0001185(100%) |
Table 5 The secondary metabolite ofbiocontrol bacterium XC-29 predicted by antiSMASH
| 基因簇 | 次级代谢产物类型 | 基因数量 | 相似基因簇 | 相似度 |
|---|---|---|---|---|
| 簇1 | 非核糖体肽合成酶 | 43 | 地衣素 | BGC0000381(92%) |
| 簇2 | 非核糖体肽合成酶、Ⅰ型聚酮类化合物 | 48 | 双效菌素 | BGC0001059(18%) |
| 簇3 | 包含Rev响应元件 | 24 | 铁载体二异羟肟酸酯 | - |
| 簇4 | 萜类化合物、镍-铁载体 | 27 | - | BGC0002683(60%) |
| 簇5 | β-内酯 | 24 | 丰原素 | BGC0001095(53%) |
| 簇6 | 萜类化合物、镍-铁载体 | 21 | - | - |
| 簇7 | Ⅲ型聚酮类化合物 | 47 | - | - |
| 簇8 | 核糖体合成和翻译后修饰肽类似物 | 15 | - | - |
| 簇9 | β-内酯 | 33 | - | - |
| 簇10 | 其他 | 46 | 杆菌溶素 | BGC0001184(85%) |
| 簇11 | 非核糖体肽-载体、非核糖体肽合成酶 | 39 | 嗜铁素bacilibactin(E/F) | BGC0002695(80%) |
| 嗜铁素bacilibactin | BGC0000309(100%) | |||
| 嗜铁素paenibactin | BGC0000401(100%) | |||
| 嗜铁素bacilibactin | BGC0001185(100%) |
| 1 | 赵亚南,黄大野,杨丹,等.烟草黑胫病研究进展[J].湖北农业科学,2022,61(S1):25-28+66. |
| ZHAO Y N, HUANG D Y, YANG D,et al.. Research progress of tobacco black shank disease[J]. Hubei Agric. Sci., 2022, 61(S1): 25-28+66. | |
| 2 | NIU B, WANG W, YUAN Z, et al.. Microbial interactions within multiple-strain biological control agents impact soil-borne plant disease[J/OL]. Front. Microbiol., 2020, 11: 585404[2024-07-25]. . |
| 3 | 蒲欣,吴茂华,刘锋,等.芽孢杆菌对玉米真菌病害生物防治效果的研究进展[J].江苏农业科学,2024,52(4):23-30. |
| PU X, WU M H, LIU F, et al.. Research progress on biological control effect of Bacillus on corn fungal diseases[J]. Jiangsu Agric. Sci., 2024, 52(4): 23-30. | |
| 4 | 刘开辉,刘月,陈妮,等.芽孢杆菌A-1的鉴定及其抗病促生作用研究[J].陕西科技大学学报,2023,41(5):64-69+86. |
| LIU K H, LIU Y, CHEN N, et al.. Study on identification of Bacillus sp. A-1 and its antipathogenic and plant growth-promoting capability[J]. J. Shaanxi Univ. Sci. Technol., 2023, 41(5): 64-69+86. | |
| 5 | 马乔女,李心悦,顾欣,等.芽孢杆菌抗真菌肽的研究进展[J].中国植保导刊,2023,43(5):17-24. |
| MA Q N, LI X Y, GU X, et al.. Research progress of Bacillus antifungal peptides[J]. China Plant Prot., 2023, 43(5): 17-24. | |
| 6 | HUSSAIN S, TAI B, ALI M, et al.. Antifungal potential of lipopeptides produced by the Bacillus siamensis Sh420 strain against Fusarium graminearum [J/OL]. Microbiol. Spectr., 2024, 12(4): e0400823[2024-07-25]. . |
| 7 | 王伟宸,赵进,黄薇颐,等.芽胞杆菌代谢产物防治三种常见植物病原真菌的研究进展[J].生物技术通报,2023,39(3):59-68. |
| WANG W C, ZHAO J, HUANG W Y, et al.. Research progress in metabolites produced by Bacillus against three common plant pathogenic fungi[J]. Biotechnol. Bull., 2023, 39(3): 59-68. | |
| 8 | 王冲,李倩,肖红英,等.贝莱斯芽孢杆菌Vel-HNGD-F2产抗菌物质发酵条件优化及抗菌特性研究[J].河南工业大学学报(自然科学),2024,45(1):73-80. |
| WANG C, LI Q, XIAO H Y, et al.. Optimization of fermentation conditions and antifungal properties of Bacillus velezensis VelHNGD-F2[J]. J. Henan Uni. Technol., 2024, 45(1): 73-80. | |
| 9 | XUE J, SUN L, XU H, et al.. Bacillus atrophaeus NX-12 utilizes exosmotic glycerol from Fusarium oxysporum f. sp. cucumerinum for fengycin production[J]. J. Agric. Food Chem., 2023, 71(28): 10565-10574. |
| 10 | NGALIMAT M S, YAHAYA R S R, BAHARUDIN M M A, et al.. A review on the biotechnological applications of the operational group Bacillus amyloliquefaciens [J/OL]. Microorganisms, 2021, 9(3): 614[2024-07-25]. . |
| 11 | 王怡凡,刘巍,朱其立,等.马铃薯早疫病拮抗细菌WK-1的筛选鉴定及其生物学特性分析[J].西南农业学报,2022,35(4):855-863. |
| WANG Y F, LIU W, ZHU Q L, et al.. Screening and identification of potato early blight antagonistic bacteria WK-1 and analysis of its biological characteristics[J]. Southwest China J. Agric. Sci., 2022, 35(4): 855-863. | |
| 12 | 贾孟媛,王越洋,唐培培,等.烟草黑胫病生防菌的筛选鉴定及其防效[J].湖南农业大学学报(自然科学),2023,49(3):329-334. |
| JIA M Y, WANG Y Y, TANG P P, et al.. Screening and identification of biocontrol bacteria for tobacco black shank disease and evaluation of the control effect[J]. J. Hunan Agric. Univ., 2023, 49(3): 329-334. | |
| 13 | 李苗苗,王晓强,王东坤,等.生防菌复配对烟草黑胫病的防治效果研究[J].中国烟草科学,2020,41(2):32-38. |
| LI M M, WANG X Q, WANG D K, et al.. Effect of biocontrol agents mixture on control of tobacco black shank[J]. Chin. Tob. Sci., 2020, 41(2): 32-38. | |
| 14 | KUMAR S, STECHER G, TAMURA K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets[J]. Mol. Biol. Evol., 2016, 33(7): 1870-1874. |
| 15 | CANTAREL B L, COUTINHO P M, RANCUREL C, et al.. The carbohydrate-active enzymes database (CAZy): an expert resource for glycogenomics[J]. Nucleic Acids Res., 2009, 37(S1): 233-238. |
| 16 | ASHBURNER M, BALL C A, BLAKE J A, et al.. Gene ontology: tool for the unification of biology. The gene ontology Consortium[J]. Cell Death Discov., 2000, 25(1): 25-29. |
| 17 | KANEHISA M, GOTO S, HATTORI M, et al.. From genomics to chemical genomics: new developments in KEGG[J]. Nucleic Acids Res., 2006, 34(S1): 354-357. |
| 18 | TATUSOV R L, FEDOROVA N D, JACKSON J D, et al.. The COG database: an updated version includes eukaryotes[J/OL]. BMC Bioinform., 2003, 4: 41[2024-07-25]. . |
| 19 | MEDEMA M H, BLIN K, CIMERMANCIC P, et al.. AntiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences[J]. Nucleic Acids Res., 2011, 39(S2): 339-346. |
| 20 | RICHTER M, ROSSELLÓ-MÓRA R, OLIVER GLÖCKNER F, et al.. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison[J]. Bioinformatics, 2016, 32(6): 929-931. |
| 21 | 濮永瑜,包玲凤,何翔,等.烟草青枯病和黑胫病拮抗细菌的筛选、鉴定及防效研究[J].中国农学通报,2022,38(7):116-123. |
| PU Y Y, BAO L F, HE X, et al.. Screening, identification and control efficacy of antagonistic bacteria against Ralstonia solanacearum and Phytophthora parasitica [J]. Chin. Agric. Sci. Bull., 2022, 38(7): 116-123. | |
| 22 | 何明川,施春兰,魏聪聪,等.烟草黑胫病拮抗细菌的分离、鉴定及发酵条件优化[J].南方农业学报,2022,53(6):1604-1615. |
| HE M C, SHI C L, WEI C C, et al.. Isolation, identification and optimization of fermentation conditions of antagonistic bacteria against tobacco black shank[J]. J. South. Agric., 2022, 53(6): 1604-1615. | |
| 23 | 谢强,夏建华,徐传涛,等.巨大芽胞杆菌(Bacillus megaterium,Bm)的抑菌活性及定殖规律分析[J].烟草科技,2022,55(10):19-25. |
| XIE Q, XIA J H, XU C T, et al.. Antibacterial activity of Bacillus megaterium strain Bm and its colonization laws[J]. Tob. Sci. Technol., 2022, 55(10): 19-25. | |
| 24 | 李小杰,李成军,刘红彦,等.烟草疫霉菌拮抗细菌的筛选鉴定及发酵条件优化[J].中国烟草科学,2019,40(1): 68-74. |
| LI X J, LI C J, LIU H Yet al.. Screening and fermentation condition optimization for antagonistic bacteria to Phytophthora nicotianae [J]. Chin. Tob. Sci., 2019, 40(1): 68-74. | |
| 25 | 李颖颖,康业斌,李成军,等.3种拮抗烟草疫霉及产IAA内生细菌的分离鉴定[J].江苏农业科学,2023,51(18):107-114. |
| LI Y Y, KANG Y B, LI C J, et al.. Isolation and identification of three endophytic bacteria antagonizing Phytophthora nicotianae and producing IAA[J]. Jiangsu Agric. Sci., 2023, 51(18): 107-114. | |
| 26 | VOLYNCHIKOVA E, KIM K D. Biological control of oomycete soilborne diseases caused by Phytophthora capsici, Phytophthora infestans, and Phytophthora nicotianae in solanaceous crops[J]. Mycobiology, 2022, 50(5): 269-293. |
| 27 | YANG J, YUE H R, PAN L Y, et al.. Fungal strain improvement for efficient cellulase production and lignocellulosic biorefinery: current status and future prospects[J/OL]. Bioresour. Technol., 2023, 385: 129449[2024-07-25]. . |
| 28 | 刘丽阳,胡彦波,王西,等.酶在植物多糖研究中的应用进展[J].食品研究与开发,2024,45(8):217-224. |
| LIU L Y, HU Y B, WANG X, et al.. Research progress on the application of enzymes in plant polysaccharides[J]. Food Res. Dev., 2024, 45(8): 217-224. | |
| 29 | CHÁVEZ-RAMÍREZ B, RODRÍGUEZ-VELÁZQUEZ N D, MONDRAGÓN-TALONIA C M, et al.. Paenibacillus polymyxa NMA1017 as a potential biocontrol agent of Phytophthora tropicalis, causal agent of cacao black pod rot in Chiapas, Mexico [J]. Antonie Van Leeuwenhoek, 2021, 114(1): 55-68. |
| 30 | WANG Y, LIANG J, ZHANG C, et al.. Bacillus megaterium WL-3 lipopeptides collaborate against Phytophthora infestans to control potato late blight and promote potato plant growth[J/OL]. Front. Microbiol., 2020, 11: 1602[2024-07-25]. . |
| 31 | HAN X, SHEN D, XIONG Q, et al.. The plant-beneficial rhizobacterium Bacillus velezensis FZB42 controls the soybean pathogen Phytophthora sojae due to bacilysin production[J/OL]. Appl. Environ. Microbiol., 2021, 87(23): e0160121[2024-07-25]. . |
| 32 | GUO D, YUAN C, LUO Y, et al.. Biocontrol of tobacco black shank disease (Phytophthora nicotianae) by Bacillus velezensis Ba168[J/OL]. Pestic. Biochem. Physiol., 2020, 165: 104523[2024-07-25]. . |
| [1] | Kai ZOU. Research and Application of Plant Hairy Root [J]. Current Biotechnology, 2024, 14(3): 341-348. |
| [2] | Liwen WANG, Jiangkun WANG, Bingbing WANG, Jianhong XU, Jianrong SHI, Xin LIU. Roles of Fusarium Toxins in Plant-pathogen Interaction [J]. Current Biotechnology, 2024, 14(2): 182-188. |
| [3] | Qiaoli CHEN, Jie HUANG, Senyu CHEN, Shaoting PAN, Lingzhi TANG, Xuan HONG. Research Progress on Secondary Metabolites of Marine Streptomyces [J]. Current Biotechnology, 2023, 13(6): 844-852. |
| [4] | Yingrui GAO, Fuzhong KANG, Tiejian MENG, Kefei LIU, Tiaotiao WANG, Jinyan CHEN, Tong SUN. Safety Evaluation of Clostridium butyricum Based on Whole Genome Sequencing [J]. Current Biotechnology, 2023, 13(5): 755-759. |
| [5] | Kainan SONG, Linan XIE, Yuquan XU. Progress of Fungal Secondary Metabolites with Potential Herbicidal Activity [J]. Current Biotechnology, 2023, 13(2): 181-194. |
| [6] | Yang YANG, Fenglin WANG, De LIU, Yuanyuan LUO, Jianhua ZHU. Research Progress of CRISPR⁃Cas9 Technology on the Production of Plant Secondary Metabolites [J]. Current Biotechnology, 2022, 12(6): 806-816. |
| [7] | LI Meng-ying, YAN Pei-sheng*, GAO Xiu-jun, SUN Xiao-lei. Progress on Diversity and Secondary Metabolites Biological Activity of Deep-sea Fungi [J]. Curr. Biotech., 2015, 5(3): 170-175. |
| [8] | WU Shui-long, JIANG Li-ming*. Recent Advances in Mangrove Actinomycetes [J]. journal1, 2012, 2(5): 335-340. |
| [9] | HUANG Yan, HU Jian-wei, ZHU Hong-hui. Research Progress in Macrocyclic Compounds of Secondary Metabolites from Myxobacteria [J]. Current Biotechnology, 2012, 2(4): 263-269. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||