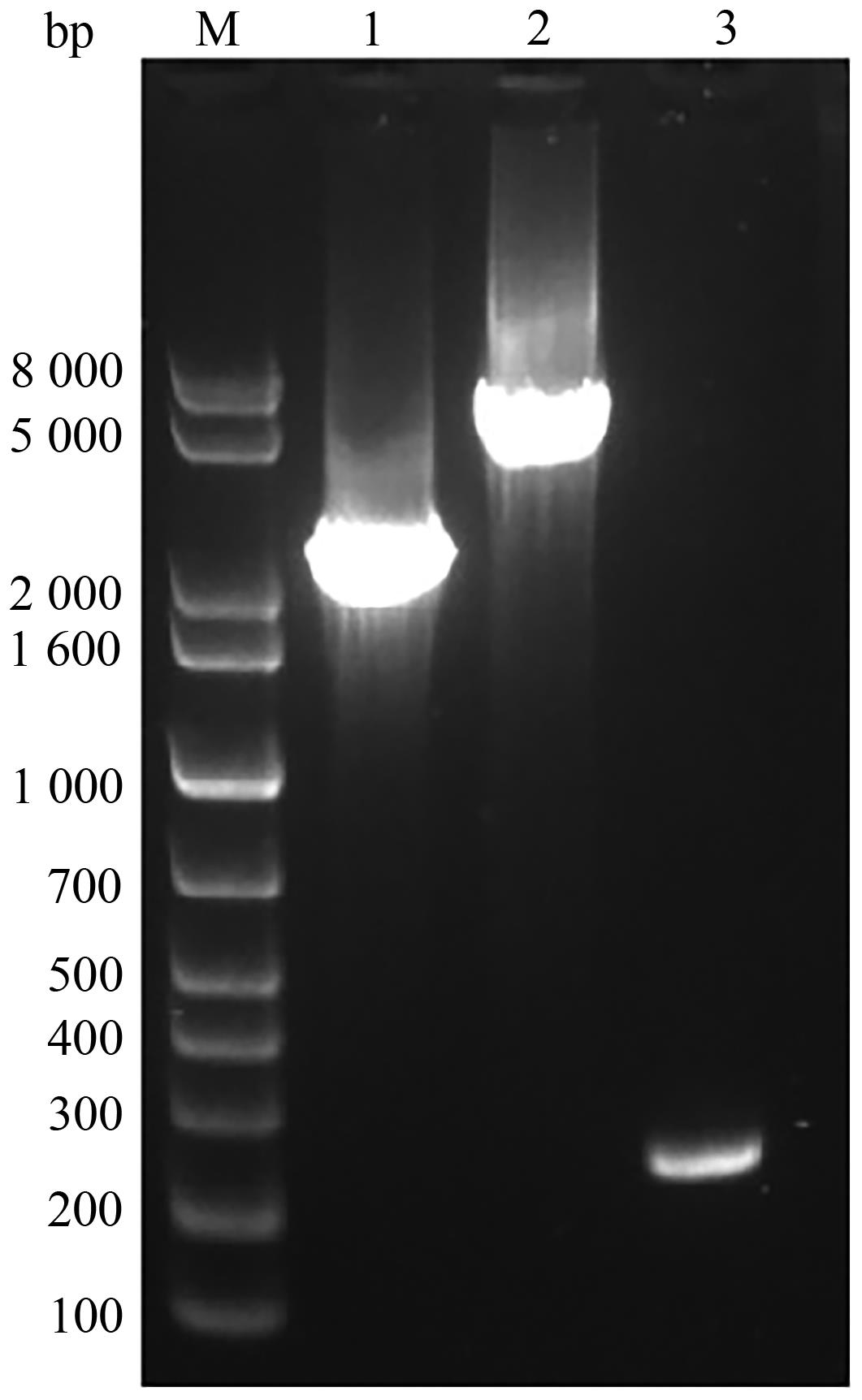
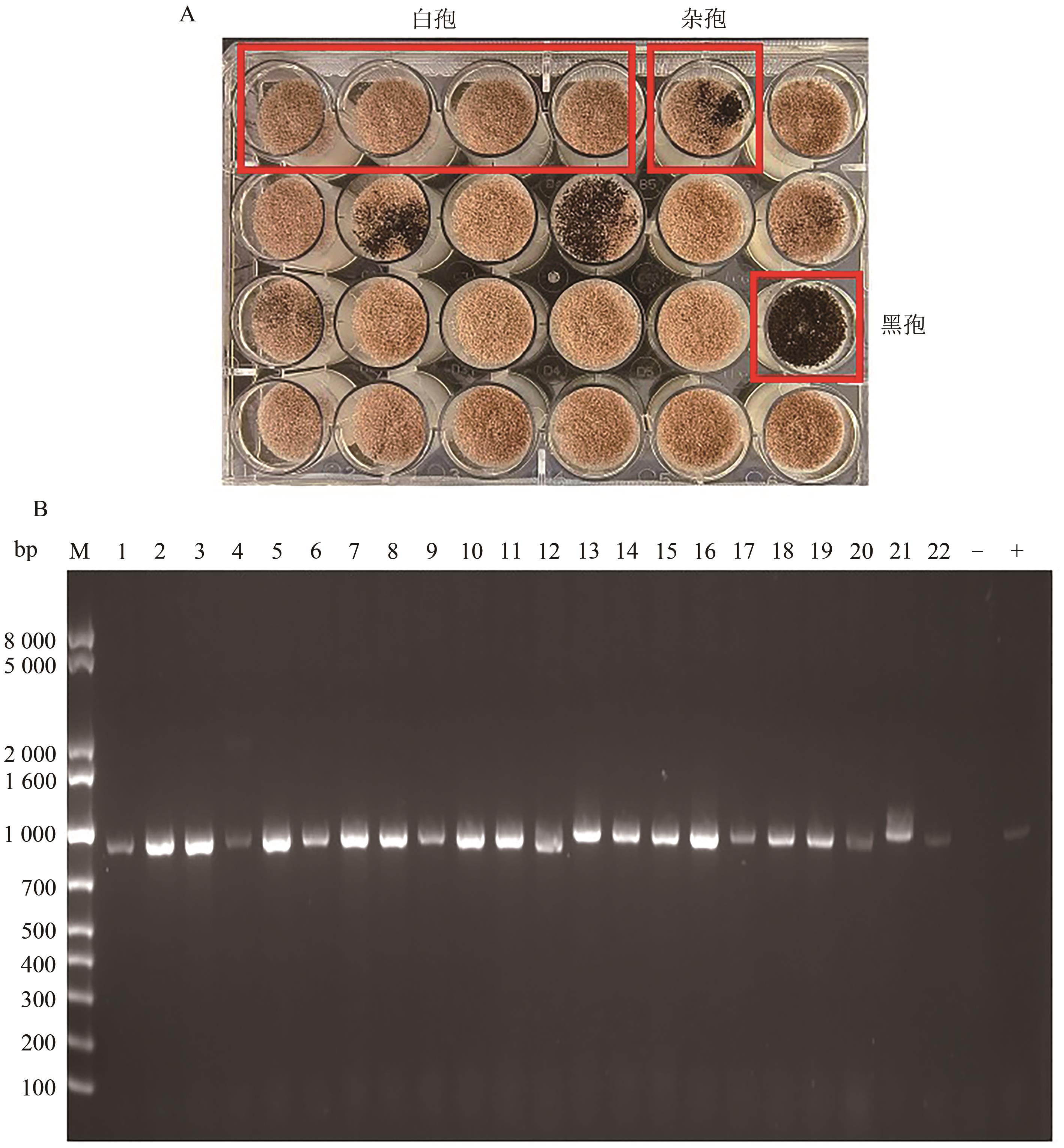
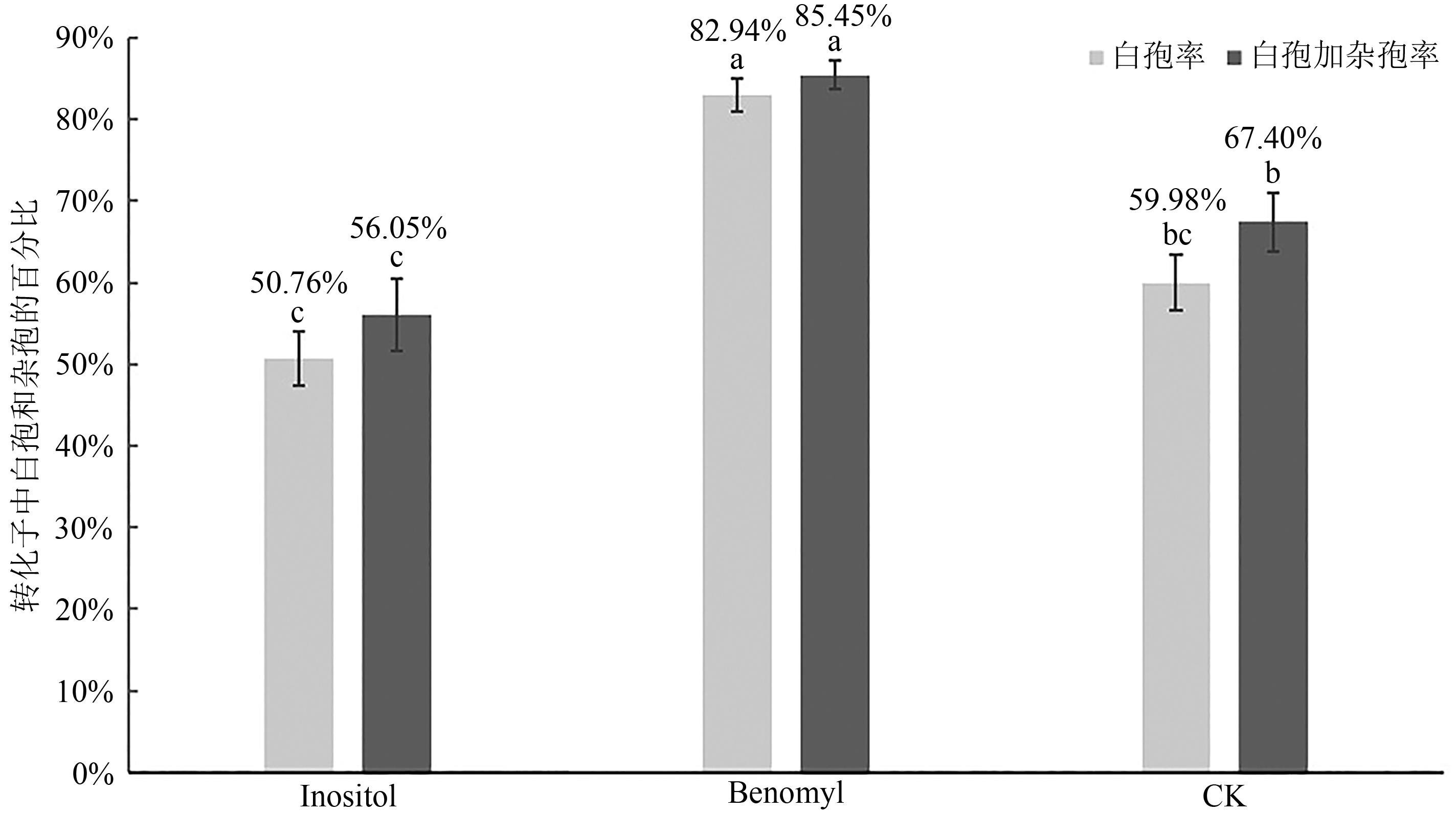
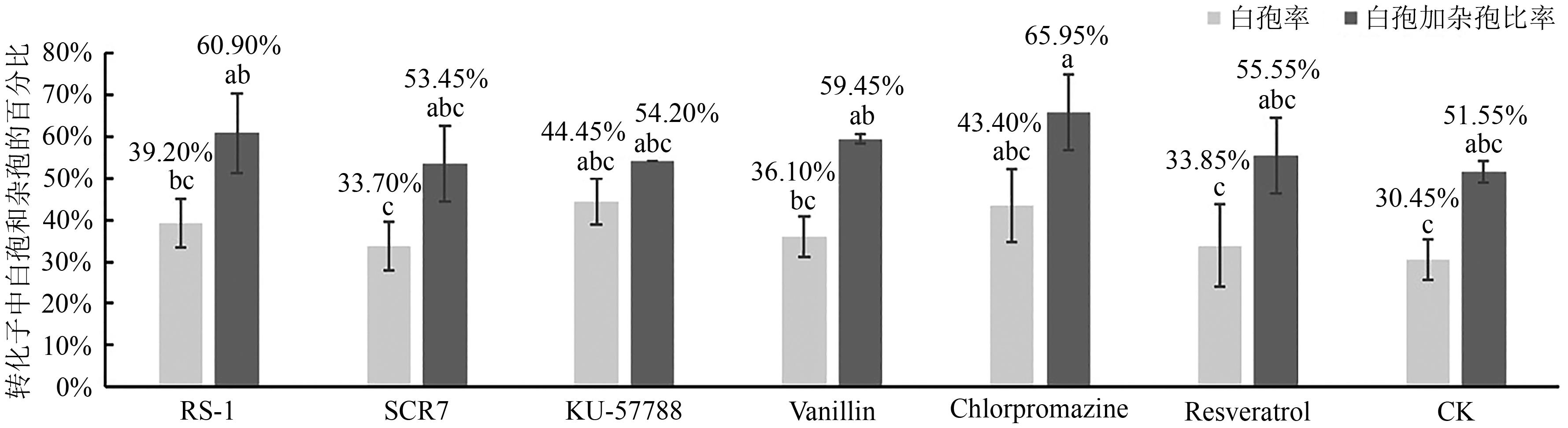
| 1 |
PÉREZ-CANTERO A, MARTIN-VICENTE A, GUARRO J, et al.. Analysis of the cyp51 genes contribution to azole resistance in Aspergillus section Nigri with the CRISPR-Cas9 technique[J]. Antimicrob. Agents Chemother., 2023, 65(5):1996- 1920.
|
| 2 |
MIRHENDI H, ZAREI F, MOTAMEDI M, et al.. Aspergillus tubingensis and Aspergillus niger as the dominant black Aspergillus, use of simple PCR-RFLP for preliminary differentiation[J]. J. Mycol. Med., 2016, 26(1): 9-16.
|
| 3 |
JIN F J, WANG B T, WANG Z D, et al.. CRISPR/Cas9-based genome editing and its application in Aspergillus species[J/OL]. J. Fungi Basel., 2022, 8(5): 467[2024-06-13]. .
|
| 4 |
YOSHIOKA I, KIRIMURA K. Rapid and marker-free gene replacement in citric acid-producing Aspergillus tubingensis (A. niger) WU-2223L by the CRISPR/Cas9 system-based genome editing technique using DNA fragments encoding sgRNAs[J]. J. Biosci. Bioeng., 2021, 131(6): 579-588.
|
| 5 |
LIU M, REHMAN S, TANG X, et al.. Methodologies for improving HDR efficiency[J/OL]. Genetics, 2018, 9: 691[2024-06-13]. .
|
| 6 |
GARRIGUES S, PENG M, KUN R S, et al.. Non-homologous end-joining-deficient filamentous fungal strains mitigate the impact of off-target mutations during the application of CRISPR/Cas9[J/OL]. mBio, 2023, 14(4): e0066823[2024-06-13]. .
|
| 7 |
高育青,张豪杰,张丹凤,等.米曲霉RIB40高效同源重组和尿苷/尿嘧啶营养缺陷型菌株的构建[J].食品工业科技,2023,44(1):200-207.
|
|
GAO Y Q, ZHANG H J, ZHANG D F, et al.. Construction of A high-efficiency homologous recombination and uridine/uracil auxotroph strain of Aspergillus oryzae RIB40[J]. Sci. Technol. Food Ind., 2023, 44(1): 200-207.
|
| 8 |
GANDÍA M, XU S, FONT C, et al.. Disruption of Ku70 involved in non-homologous end-joining facilitates homologous recombination but increases temperature sensitivity in the phytopathogenic fungus Penicillium digitatum [J]. Fungal. Biol., 2016, 120(3): 317-323.
|
| 9 |
FANG Z, ZHANG Y, CAI M, et al.. Improved gene targeting frequency in marine-derived filamentous fungus Aspergillus glaucus by disrupting ligD[J]. J. Appl. Genet., 2012, 53(3): 355-362.
|
| 10 |
YU C, LIU Y, MA T, et al.. Small molecules enhance CRISPR genome editing in pluripotent stem cells[J]. Cell Stem Cell, 2015, 16(2): 142-147.
|
| 11 |
MA X, CHEN X, JIN Y, et al.. Small molecules promote CRISPR-Cpf1-mediated genome editing in human pluripotent stem cells[J/OL]. Nat. Commun., 2018, 9: 1303[2024-06-13]. .
|
| 12 |
YEH C D, RICHARDSON C D, CORN J E. Advances in genome editing through control of DNA repair pathways[J]. Nat. Cell Biol., 2019, 21(12): 1468-1478.
|
| 13 |
ZOU G, XIAO M, CHAI S, et al.. Efficient genome editing in filamentous fungi via an improved CRISPR-Cas9 ribonucleoprotein method facilitated by chemical reagents[J]. Microb. Biotechnol., 2021, 14(6): 2343-2355.
|
| 14 |
SONG J, YANG D, XU J, et al.. RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency[J/OL]. Nat. Commun., 2016, 7: 10548[2024-06-13]. .
|
| 15 |
CHU V T, WEBER T, WEFERS B, et al.. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells[J]. Nat. Biotechnol., 2015, 33: 543-548.
|
| 16 |
GAVANDE N S, VANDERVERE-CAROZZA P S, HINSHAW H D, et al.. DNA repair targeted therapy: the past or future of cancer treatment?[J]. Pharmacol. Ther., 2016, 160: 65-83.
|
| 17 |
DING Y, WANG K F, WANG W J, et al.. Increasing the homologous recombination efficiency of eukaryotic microorganisms for enhanced genome engineering[J]. Appl. Microbiol. Biotechnol., 2019, 103(11): 4313-4324.
|
| 18 |
SINHA S, CHATTERJEE S, PAUL S, et al.. Olaparib enhances the Resveratrol-mediated apoptosis in breast cancer cells by inhibiting the homologous recombination repair pathway[J/OL]. Exp. Cell Res., 2022, 420(1): 113338[2024-06-13]. .
|
| 19 |
高伟欣,黄火清,赵晶,等.应用于基因编辑的核糖核蛋白复合体的构建与活性验证[J].生物技术通报,2022,38(8):60-68.
|
|
GAO W X, HUANG H Q, ZHAO J, et al.. Construction and activity verification of ribonucleoprotein complex for gene editing[J]. Biotechnol. Bull., 2022, 38(8): 60-68.
|
| 20 |
BISCHOFF N, WIMBERGER S, MARESCA M, et al.. Improving precise CRISPR genome editing by small molecules: is there a magic potion?[J/OL]. Cells, 2020, 9(5): E1318[2024-06-13]. .
|
 ), Wenlong LIU2, Xiaoqing LIU1, Bin YAO1, Huoqing HUANG1, Haomeng YANG1(
), Wenlong LIU2, Xiaoqing LIU1, Bin YAO1, Huoqing HUANG1, Haomeng YANG1( )
)
 ), 刘文龙2, 刘晓青1, 姚斌1, 黄火清1, 杨浩萌1(
), 刘文龙2, 刘晓青1, 姚斌1, 黄火清1, 杨浩萌1( )
)