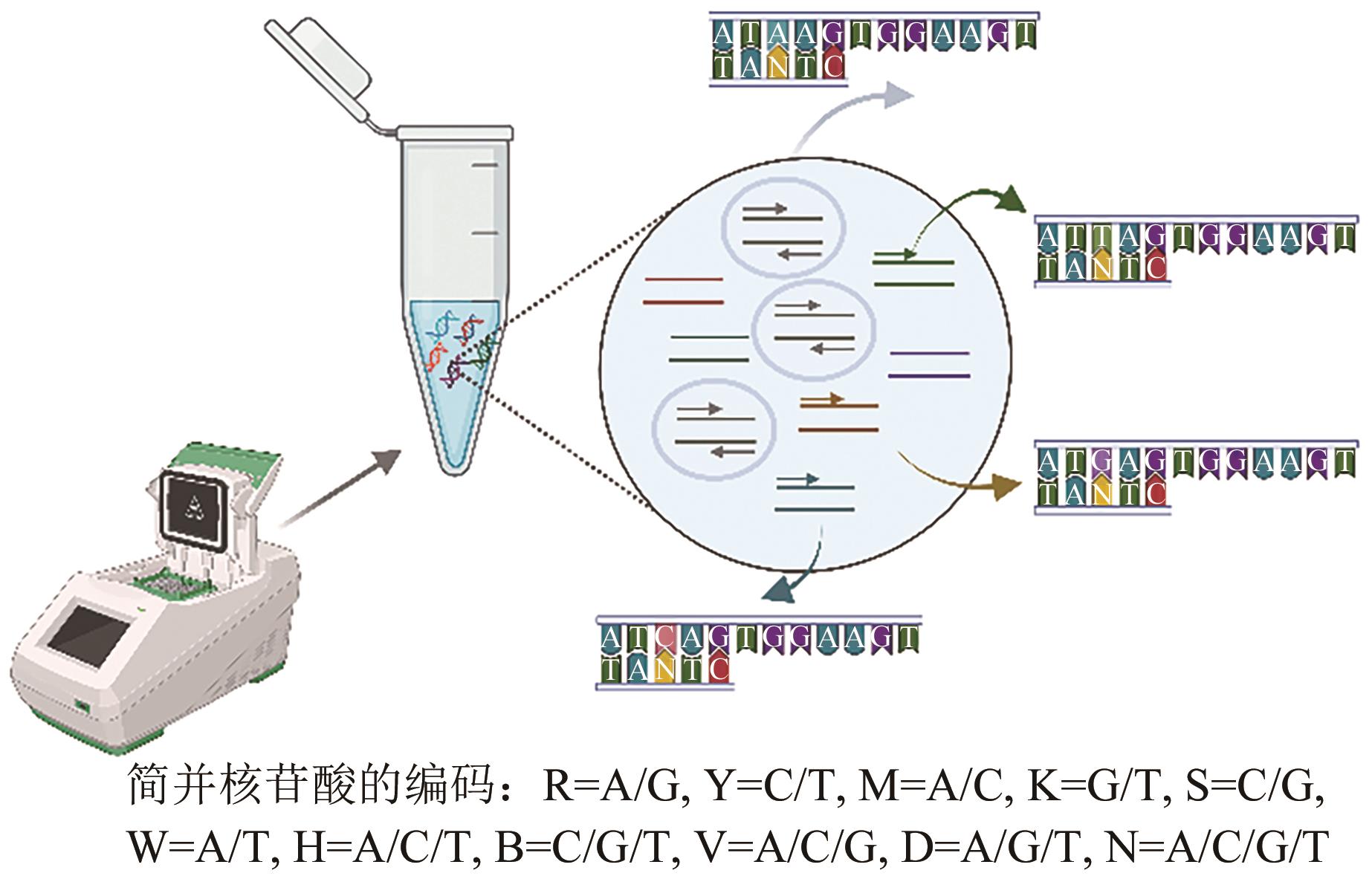
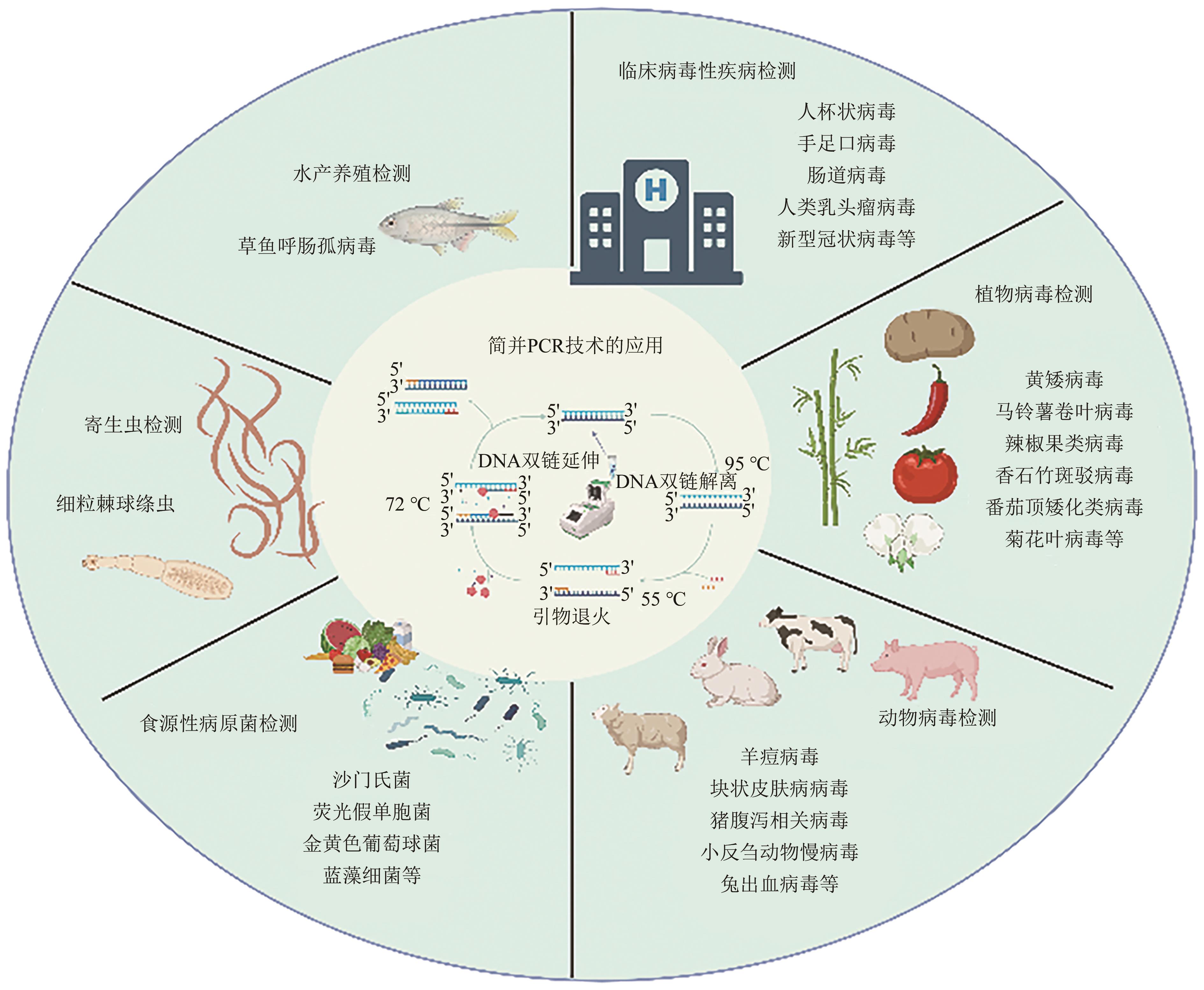
Current Biotechnology ›› 2024, Vol. 14 ›› Issue (4): 545-554.DOI: 10.19586/j.2095-2341.2024.0061
• Reviews • Previous Articles Next Articles
Application of Degenerate PCR Technique in Disease Detection
Min LIU1( ), Huai CHENG1, Huanchang CAI1, Hewei ZHANG2(
), Huai CHENG1, Huanchang CAI1, Hewei ZHANG2( ), Jingqiang REN1(
), Jingqiang REN1( )
)
- 1.Institute of Virology,Wenzhou University,Zhejiang Wenzhou 325035,China
2.College of Food and Drug,Luoyang Vocational and Technical College,Henan Luoyang 471400,China
-
Received:2024-03-26Accepted:2024-05-28Online:2024-07-25Published:2024-08-07 -
Contact:Hewei ZHANG,Jingqiang REN
简并PCR技术在疾病检测中的应用
刘敏1( ), 程淮1, 蔡欢嫦1, 张贺伟2(
), 程淮1, 蔡欢嫦1, 张贺伟2( ), 任静强1(
), 任静强1( )
)
- 1.温州大学病毒学研究所,浙江 温州 325035
2.洛阳职业技术学院食品与药品学院,河南 洛阳 471400
-
通讯作者:张贺伟,任静强 -
作者简介:刘敏 E-mail: min1011@163.com -
基金资助:温州大学引进人才科研启动项目(QD2023014);温州市基础性科研资助项目(20231107)
CLC Number:
Cite this article
Min LIU, Huai CHENG, Huanchang CAI, Hewei ZHANG, Jingqiang REN. Application of Degenerate PCR Technique in Disease Detection[J]. Current Biotechnology, 2024, 14(4): 545-554.
刘敏, 程淮, 蔡欢嫦, 张贺伟, 任静强. 简并PCR技术在疾病检测中的应用[J]. 生物技术进展, 2024, 14(4): 545-554.
share this article
| 1 | 钱佳婕,黄迪,徐颖华,等.食源性致病微生物检测技术研究进展[J].食品安全质量检测学报,2021,12(12):4775-4785. |
| QIAN J J, HUANG D, XU Y H, et al.. Research progress of detection technologies for foodborne pathogens[J]. J. Food Saf. Qual., 2021, 12(12): 4775-4785. | |
| 2 | MARKOULATOS P, SIAFAKAS N, MONCANY M. Multiplex polymerase chain reaction: a practical approach[J]. J. Clin. Lab. Anal., 2002, 16(1): 47-51. |
| 3 | RAJALAKSHMI S. Different types of PCR techniques and its application[J].Int.J.Pharm.Chem. Biol. Sci., 2017, 7(3): 285-292. |
| 4 | 史兆兴,王恒樑,苏国富,等. 简并PCR及其应用[J].生物技术通讯,2004,15(2):172-175. |
| SHI Z X, WANG H L, SU G F,et al..简并PCR及其应用[J].Lett. Biotechnol.,2004,15(2):172-175. | |
| 5 | JORDAN B, CHAREST A, DOWD J F, et al.. Genome complexity reduction for SNP genotyping analysis[J]. Proc. Natl. Acad. Sci. USA, 2002, 99(5): 2942-2947. |
| 6 | NAYAK S N, BALAJI J, UPADHYAYA H D, et al.. Isolation and sequence analysis of DREB2A homologues in three cereal and two legume species[J]. Plant Sci., 2009, 177(5): 460-467. |
| 7 | GUYADER F L, ESTES M K, HARDY M E, et al.. Evaluation of a degenerate primer for the PCR detection of human caliciviruses[J]. Arch. Virol., 1996, 141(11): 2225-2235. |
| 8 | CHANG T J, HSIA C Y, CHAU G Y, et al.. Characterization of androgen receptor complex associated protein (ARCAP) in hepatocellular carcinoma and liver[J]. J. Chin. Med. Assoc., 2021, 84(12): 1100-1108. |
| 9 | 杨怀涛,杨秀英.检测HPV的新策略:简并引物PCR[J].国外医学·临床生物化学与检验学分册,1994,15(2):71-73. |
| YANG H T, YANG X Y. A new strategy for HPV detection-degenerate primer PCR[J]. Foreign Med. Sci., 1994, 15(2): 71-73. | |
| 10 | 侯林,李楠,伊男,等.中国卤虫早期胚胎发育相关基因的研究: Orthodenticle基因克隆中简并引物的设计和优化[J].辽宁师范大学学报(自然科学版),2009,32(1):98-101. |
| HOU L, LI N, YI N, et al.. A study on the related genes of early embryonic development of Artemia sinica: degenerate PCR primer design and improvement for Artemia sinica Orthodenticle clone[J]. J. Liaoning Norm. Univ. Nat. Sci. Ed., 2009, 32(1): 98-101. | |
| 11 | NAQIB A, JEON T, KUNSTMAN K, et al.. PCR effects of melting temperature adjustment of individual primers in degenerate primer pools [J/OL]. PeerJ, 2019, 7: e6570[2024-0306]. . |
| 12 | SAS M A, VINA-RODRIGUEZ A, MERTENS M, et al.. A one-step multiplex real-time RT-PCR for the universal detection of all currently known CCHFV genotypes[J]. J. Virol. Meth., 2018, 255: 38-43. |
| 13 | 王依,胡丹,钟璟皓,等.基于序列非依赖性扩增技术发现未知病毒的研究进展[J].生物技术通讯,2016,27(4):576-581. |
| WANG Y, HU D, ZHONG J H, et al.. Discover unknown viruses based metagenomics sequence-independent amplification[J]. Lett. Biotechnol., 2016, 27(4): 576-581. | |
| 14 | CHOI S, KIM K W, KU K B, et al.. Human alphacoronavirus universal primers for genome amplification and sequencing[J/OL]. Front. Microbiol., 2022, 13: 789665[2024-06-17]. . |
| 15 | 王少峡,陈丽媛,张竞秋,等.利用生物信息学资源设计简并引物[J].天津师范大学学报(自然科学版),2006,26(2):26-29. |
| WANG S X, CHEN L Y, ZHANG J Q, et al.. Designing the degenerate primers with bioinformatics resource[J]. J. Tianjin Norm. Univ. Nat. Sci. Ed., 2006, 26(2): 26-29. | |
| 16 | LINHART C, SHAMIR R. The degenerate primer design problem: theory and applications[J]. J. Comput. Biol., 2005, 12(4): 431-456. |
| 17 | JOHN G D, PAUL K, ANTHONY W R. Virus detection using degenerate PCR primers: WO099130[P].2002-06-07. |
| 18 | RECTOR A, TACHEZY R, VAN RANST M. A sequence-independent strategy for detection and cloning of circular DNA virus genomes by using multiply primed rolling-circle amplification[J]. J. Virol., 2004, 78(10): 4993-4998. |
| 19 | HO T, TZANETAKIS I E. Development of a virus detection and discovery pipeline using next generation sequencing[J]. Virology, 2014, 471-473: 54-60. |
| 20 | 胡群,倪红霞.应用一致简并引物RT-PCR检测手足口病患者粪便肠道病毒[J].中国国境卫生检疫杂志,2017,40(4):239-241+292. |
| HU Q, NI H X. Detection and genotyping of enterovirus in feces of HFMD patients by consensus degenerate hybrid oligonucleotide primers RT-PCR[J]. Chin. J. Front. Health Quar., 2017, 40(4): 239-241+292. | |
| 21 | 郭华,丁惠,李慧娟,等.儿童病毒性脑膜炎、脑炎中肠道病毒感染的分子生物学分型检测及临床研究[J].实用临床医药杂志,2016,20(1):35-38. |
| GUO H, DING H, LI H J, et al.. Molecular biology typing and clinical research of enterovirus infection in children with viral meningitis/encephalitis[J]. J. Clin. Med. Pract., 2016, 20(1): 35-38. | |
| 22 | 吴瑶,吴亮,阴晴,等.6对通用引物检测不同型别人乳头瘤病毒效果分析[J].江苏大学学报(医学版),2019,29(5):405-409. |
| WU Y, WU L, YIN Q, et al.. Detection efficacy of 6 universal primers in different types of papillomavirus[J]. J. Jiangsu Univ. Med. Ed., 2019, 29(5): 405-409. | |
| 23 | FORSLUND O, ANTONSSON A, NORDIN P, et al.. A broad range of human papillomavirus types detected with a general PCR method suitable for analysis of cutaneous tumours and normal skin[J]. J. Gen. Virol., 1999, 80( Pt 9): 2437-2443. |
| 24 | 胡群,邹春颖,马思杰.沙粒病毒属四步法一致简并杂合寡核苷酸引物实时荧光PCR检测体系的建立[J].中国媒介生物学及控制杂志,2016,27(3):248-252. |
| HU Q, ZOU C Y, MA S J. Development of CODEHOP 4-step program realtime PCR to detect Arenaviruses in rodents[J]. Chin. J. Vector Biol. Contr., 2016, 27(3): 248-252. | |
| 25 | CHU D K W, PAN Y, CHENG S M S, et al.. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of Pneumonia[J]. Clin. Chem., 2020, 66(4): 549-555. |
| 26 | HOFFMANN E, STECH J, GUAN Y, et al.. Universal primer set for the full-length amplification of all influenza A viruses[J]. Arch. Virol., 2001, 146(12): 2275-2289. |
| 27 | HARKNESS A, MATTHEWS Q L, SMITH L, et al.. Optimization of universal primer set for the identification of all influenza A viruses using real-time PCR[J/OL]. BioRxiv, 2022, doi:10.1101/2022.08.30.505914[2024-06-15]. . |
| 28 | NANDA S, JAYAN G, VOULGAROPOULOU F, et al.. Universal virus detection by degenerate-oligonucleotide primed polymerase chain reaction of purified viral nucleic acids[J]. J. Virol. Meth., 2008, 152(1-2): 18-24. |
| 29 | 廖富荣,陈红运,沈建国,等.香石竹斑驳病毒属简并引物RT-PCR检测方法的建立[J].中国农业科学,2023,56(12):2288-2301. |
| LIAO F R, CHEN H Y, SHEN J G, et al.. Development of a degenerate primer RT-PCR assay for detection of carmovirus[J]. Sci. Agric. Sin., 2023, 56(12): 2288-2301. | |
| 30 | KUBOTA K, CHIAKI Y, YANAGISAWA H, et al.. Novel degenerate primer sets for the detection and identification of emaraviruses reveal new chrysanthemum species[J/OL]. J. Virol. Methods, 2021, 288: 113992[2024-06-17]. . |
| 31 | DIGIARO M, ELBEAINO T, MARTELLI G P. Development of degenerate and species-specific primers for the differential and simultaneous RT-PCR detection of grapevine-infecting nepoviruses of subgroups A, B and C[J]. J. Virol. Methods, 2007, 141(1): 34-40. |
| 32 | TSENG Y W, WU C F, LEE C H, et al.. Universal primers for rapid detection of six pospiviroids in Solanaceae plants using one-step reverse-transcription PCR and reverse-transcription loop-mediated isothermal amplification[J]. Plant Dis., 2021, 105(10): 2867-2872. |
| 33 | NAN W, GONG M, LU Y, et al.. A novel triplex real-time PCR assay for the differentiation of lumpy skin disease virus, goatpox virus, and sheeppox virus[J/OL]. Front. Vet. Sci., 2023, 10: 1175391[2024-06-19]. . |
| 34 | NAN P, WEN D, OPRIESSNIG T, et al.. Novel universal primer-pentaplex PCR assay based on chimeric primers for simultaneous detection of five common pig viruses associated with diarrhea[J/OL]. Mol. Cell. Probes, 2021, 58: 101747[2024-06-18]. . |
| 35 | 杨义琴.小反刍动物慢病毒实时荧光定量RT-PCR检测方法的建立和初步应用[D].新疆石河子:石河子大学,2018. |
| 36 | 王波,李桂黎,王印,等.兔出血症病毒SYBR Green Ⅰ实时荧光定量PCR检测方法的建立及初步应用[J].浙江农业学报,2016,28(3):400-405. |
| WANG B, LI G L, WANG Y, et al.. Establishment and application of SYBR Green Ⅰ real-time PCR for detection of rabbit hemorrhagic disease virus[J]. Acta Agric. Zhejiangensis, 2016, 28(3): 400-405. | |
| 37 | CHASSALEVRIS T, CHAINTOUTIS S C, APOSTOLIDI E D, et al.. A highly sensitive semi-nested real-time PCR utilizing oligospermine-conjugated degenerate primers for the detection of diverse strains of small ruminant lentiviruses[J/OL]. Mol. Cell. Probes, 2020, 51: 101528[2024-06-17]. . |
| 38 | LIN C K, HUNG C L, HSU S C, et al.. An improved PCR primer pair based on 16S rDNA for the specific detection of Salmonella serovars in food samples[J]. J. Food Prot., 2004, 67(7): 1335-1343. |
| 39 | 步雨珊,乔文君,黄冬成,等.聚合酶链式反应技术快速检测原料乳中荧光假单胞菌[J].食品安全质量检测学报,2022,13(15):4766-4772. |
| BU Y S, QIAO W J, HUANG D C, et al.. Rapid detection of Pseudomonas fluorescens in raw milk based on polymerase chain reaction assay[J]. J. Food Saf. Qual., 2022, 13(15): 4766-4772. | |
| 40 | 刘继超,陈柏儒,姜铁民,等.利用简并引物检测金黄色葡萄球菌肠毒素A、B基因[J].食品与机械,2015,31(5):59-61. |
| LIU J C, CHEN B R, JIANG T M, et al.. Research on degenerate primers for detection of Staphylococcus aureus enterotoxin A, B gene[J]. Food Mach., 2015, 31(5): 59-61. | |
| 41 | DEHAUT A, KRZEWINSKI F, GRARD T, et al.. Monitoring the freshness of fish: development of a qPCR method applied to MAP chilled whiting[J]. J. Sci. Food Agric., 2016, 96(6): 2080-2089. |
| 42 | GHAZALI N S H, RASHID N H A. Molecular identification of bacterial communities from vegetables samples as revealed by DNA sequencing of universal primer 16sRNA gene[J]. Int. J. Med. Sci., 2019, 4 (1): 19-26. |
| 43 | 李飞武,闫伟,龙丽坤,等.应用多重PCR技术筛选检测转Bt基因作物[J].现代食品科技,2014,30(5):262-266+124. |
| LI F W, YAN W, LONG L K, et al.. Screening detection of Bt genes in genetically modified crops using a multiplex PCR assay[J]. Mod. Food Sci. Technol., 2014, 30(5): 262-266+124. | |
| 44 | 李葱葱,闫伟,夏蔚,等.应用简并PCR方法检测转cry1A基因作物[J].食品科学,2018,39(14):317-322. |
| LI C C, YAN W, XIA W, et al.. Detection of genetically modified crops with cry1A gene by PCR with degenerate primers[J]. Food Sci., 2018, 39(14): 317-322. | |
| 45 | LIU W, TAO J, XUE M, et al.. A multiplex PCR method mediated by universal primers for the identification of eight meat ingredients in food products[J]. Eur. Food Res. Technol., 2019, 245(11): 2385-2392. |
| 46 | LI J, LI J, XU S, et al.. A rapid and reliable multiplex PCR assay for simultaneous detection of fourteen animal species in two tubes[J]. Food Chem., 2019, 295: 395-402. |
| 47 | 王会品,李欣,孙世珺.简并引物qPCR法检测新疆喀什地区羊肝组织中细粒棘球绦虫[J].广东医学,2022,43(1):46-50. |
| WANG H P, LI X, SUN S J. Detection of Echinococcus granulosus in sheep liver from Kashi region of Xinjiang by degenerate primer qPCR method[J]. Guangdong Med. J., 2022, 43(1): 46-50. | |
| 48 | 范玉顶,马杰,周勇,等.不同基因型草鱼呼肠孤病毒通用RT-PCR检测方法的建立及应用[J].淡水渔业,2018,48(6):9-16. |
| FAN Y D, MA J, ZHOU Y, et al.. Establishment and application of a universal RT-PCR assay for detection of different genotype grass carp reovirus[J]. Freshw. Fish., 2018, 48(6): 9-16. | |
| 49 | LIU R, NING J, JIANG Y, et al.. A method for degenerate primer design based on artificial bee colony algorithm[J/OL]. Appl. Sci., 2022, 12(10): 4992[2024-06-17]. . |
| 50 | DE ARRUDA M P, GONÇALVES E C, SCHNEIDER M P, et al.. An alternative genotyping method using dye-labeled universal primer to reduce unspecific amplifications[J]. Mol. Biol. Rep., 2010, 37(4): 2031-2036. |
| 51 | VARADARAJ K, SKINNER D M. Denaturants or cosolvents improve the specificity of PCR amplification of a G + C-rich DNA using genetically engineered DNA polymerases[J]. Gene, 1994, 140(1): 1-5. |
| 52 | JANG M, KIM S. Inhibition of non-specific amplification in loop-mediated isothermal amplification via tetramethylammonium chloride[J]. Biochip J., 2022, 16(3): 326-333. |
| 53 | KOVÁROVÁ M, DRÁBER P. New specificity and yield enhancer of polymerase chain reactions[J/OL]. Nucleic Acids Res., 2000, 28(13): E70[2024-06-19]. . |
| 54 | 朱晓静,戴忠敏.利用抑制性PCR提高兼并引物扩增效率及特异性[J].杭州师范大学学报(自然科学版),2015,14(6):581-584. |
| ZHU X J, DAI Z M. Enhance the amplification efficiency and specificity of degenerate primers with suppression PCR[J]. J. Hangzhou Norm. Univ. Nat. Sci. Ed., 2015, 14(6): 581-584. |
| No related articles found! |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||