Current Biotechnology ›› 2023, Vol. 13 ›› Issue (1): 115-123.DOI: 10.19586/j.2095-2341.2022.0158
• Articles • Previous Articles Next Articles
Effect of Different Pretreatment on the Results of High-throughput Sequencing of Microbial Community in Mouse Feces
Xin LIU1( ), Huiling HU1, Minai ZHANG2, Guozhen XUE1, Xiaorui YAN1, Xichun ZHANG1(
), Huiling HU1, Minai ZHANG2, Guozhen XUE1, Xiaorui YAN1, Xichun ZHANG1( ), Jiquan LIU1(
), Jiquan LIU1( )
)
- 1.School of Traditional Chinese Medicine and Food Engineering,Shanxi University of Traditional Chinese Medicine,Shanxi Jinzhong 030600,China
2.Customs Technical Center of Taiyuan,Taiyuan 030021,China
-
Received:2020-09-08Accepted:2020-11-30Online:2023-01-25Published:2023-02-07 -
Contact:Xichun ZHANG,Jiquan LIU
不同预处理对小鼠粪样菌群高通量测序分析结果的影响
刘昕1( ), 胡会玲1, 张敏爱2, 薛帼珍1, 闫晓睿1, 张希春1(
), 胡会玲1, 张敏爱2, 薛帼珍1, 闫晓睿1, 张希春1( ), 刘计权1(
), 刘计权1( )
)
- 1.山西中医药大学中药与食品工程学院,山西 晋中 030600
2.太原海关技术中心,太原 030021
-
通讯作者:张希春,刘计权 -
作者简介:刘昕 E-mail:1113984315@qq.com; -
基金资助:山西省自然科学基金面上项目(20210302123233);山西省现代农业产业技术体系建设专项资金(2021-11);山西中医药大学科技创新科学团队项目(2018TD-009);山西中医药大学科技创新培育计划项目(2022PY-TH-30)
CLC Number:
Cite this article
Xin LIU, Huiling HU, Minai ZHANG, Guozhen XUE, Xiaorui YAN, Xichun ZHANG, Jiquan LIU. Effect of Different Pretreatment on the Results of High-throughput Sequencing of Microbial Community in Mouse Feces[J]. Current Biotechnology, 2023, 13(1): 115-123.
刘昕, 胡会玲, 张敏爱, 薛帼珍, 闫晓睿, 张希春, 刘计权. 不同预处理对小鼠粪样菌群高通量测序分析结果的影响[J]. 生物技术进展, 2023, 13(1): 115-123.
share this article
| 样本编号 | Clean-tags | Valid-tags | Valid-tags mean length/bp | OTUs |
|---|---|---|---|---|
| S1 | 76 432 | 63 910 | 412.69 | 656 |
| S2 | 77 145 | 65 163 | 412.34 | 661 |
| S3 | 77 400 | 65 576 | 412.63 | 670 |
| P1 | 72 165 | 61 166 | 419.79 | 351 |
| P2 | 75 419 | 65 217 | 420.02 | 373 |
| P3 | 75 107 | 65 796 | 421.21 | 368 |
| Y1 | 75 933 | 64 761 | 419.98 | 442 |
| Y2 | 69 909 | 56 015 | 420.04 | 424 |
| Y3 | 77 752 | 65 465 | 419.02 | 436 |
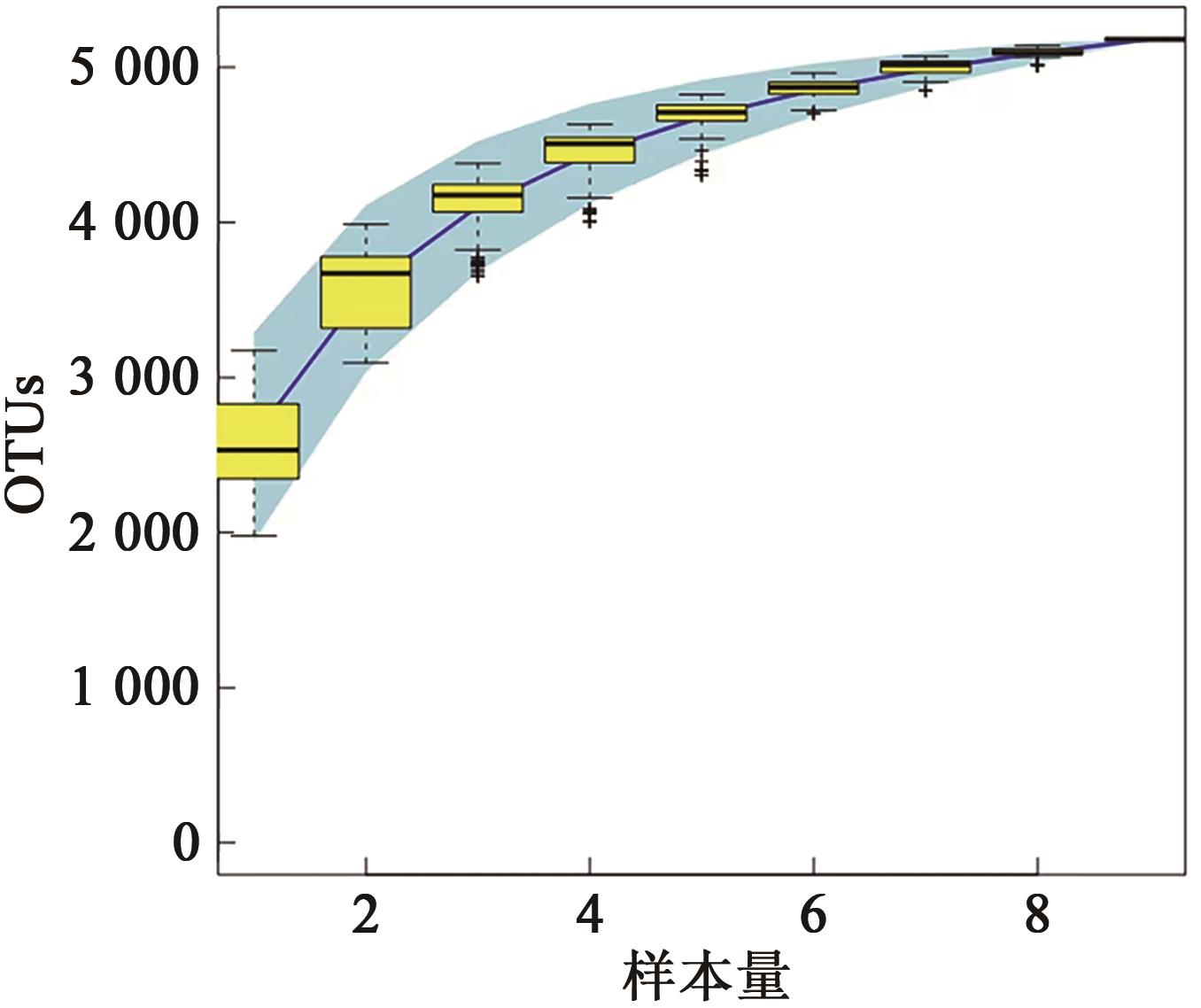
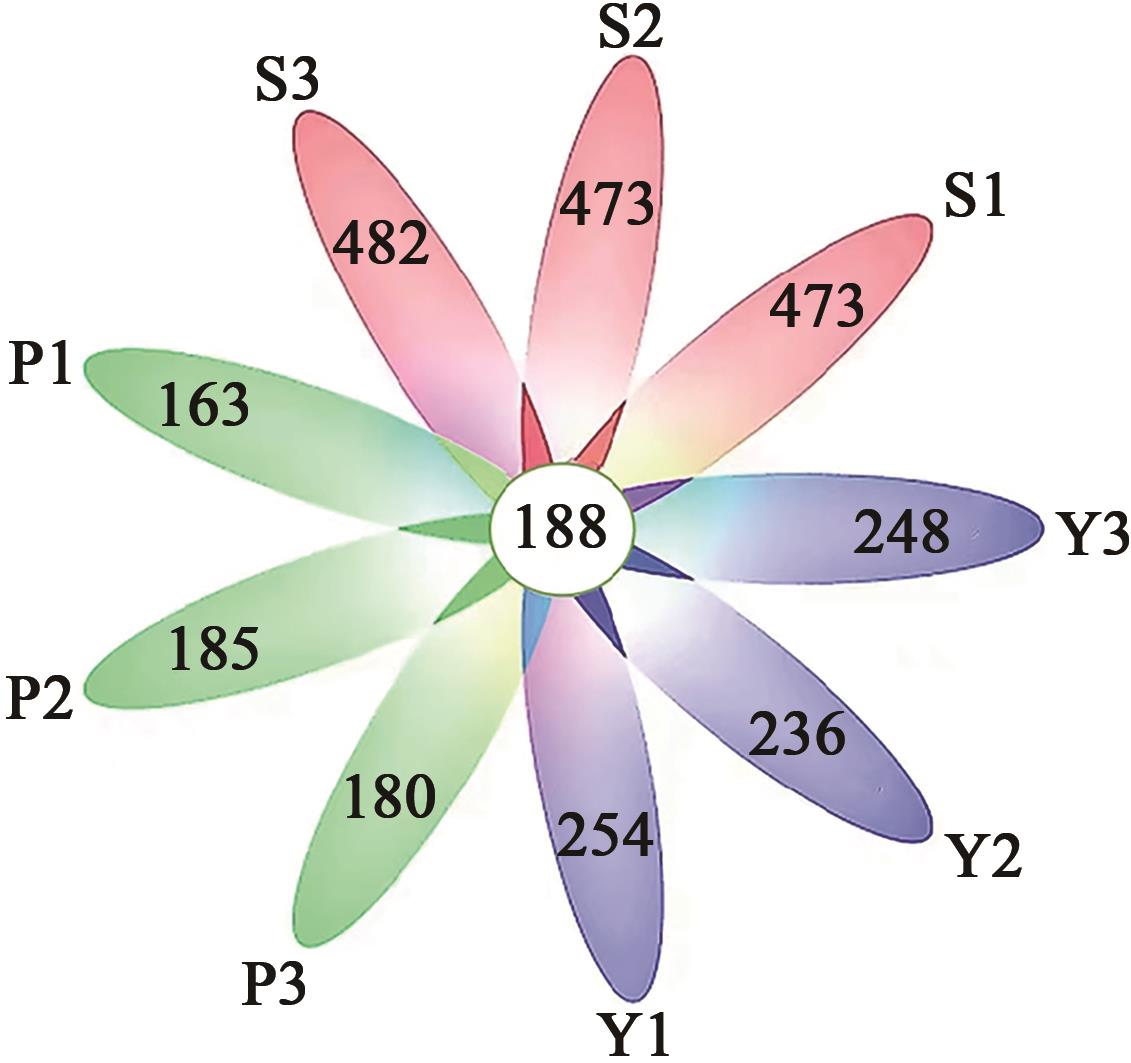
Table 1 OTUs analysis of 9 samples in 3 groups
| 样本编号 | Clean-tags | Valid-tags | Valid-tags mean length/bp | OTUs |
|---|---|---|---|---|
| S1 | 76 432 | 63 910 | 412.69 | 656 |
| S2 | 77 145 | 65 163 | 412.34 | 661 |
| S3 | 77 400 | 65 576 | 412.63 | 670 |
| P1 | 72 165 | 61 166 | 419.79 | 351 |
| P2 | 75 419 | 65 217 | 420.02 | 373 |
| P3 | 75 107 | 65 796 | 421.21 | 368 |
| Y1 | 75 933 | 64 761 | 419.98 | 442 |
| Y2 | 69 909 | 56 015 | 420.04 | 424 |
| Y3 | 77 752 | 65 465 | 419.02 | 436 |
| 组别 | Chao 1 | Observed-species | Simpson | Shannon | Goods coverage |
|---|---|---|---|---|---|
| S组 | 707.32 ± 14.090a | 650.17 ± 5.500a | 0.94 ± 0.001a | 5.59 ± 0.030a | 0.998 379a |
| P组 | 418.67 ± 10.430c | 353.07 ± 8.110c | 0.78 ± 0.010c | 3.42 ± 0.050c | 0.998 581a |
| Y组 | 496.76 ± 12.680b | 424.83 ± 3.820b | 0.85 ± 0.020b | 4.12 ± 0.050b | 0.998 225a |
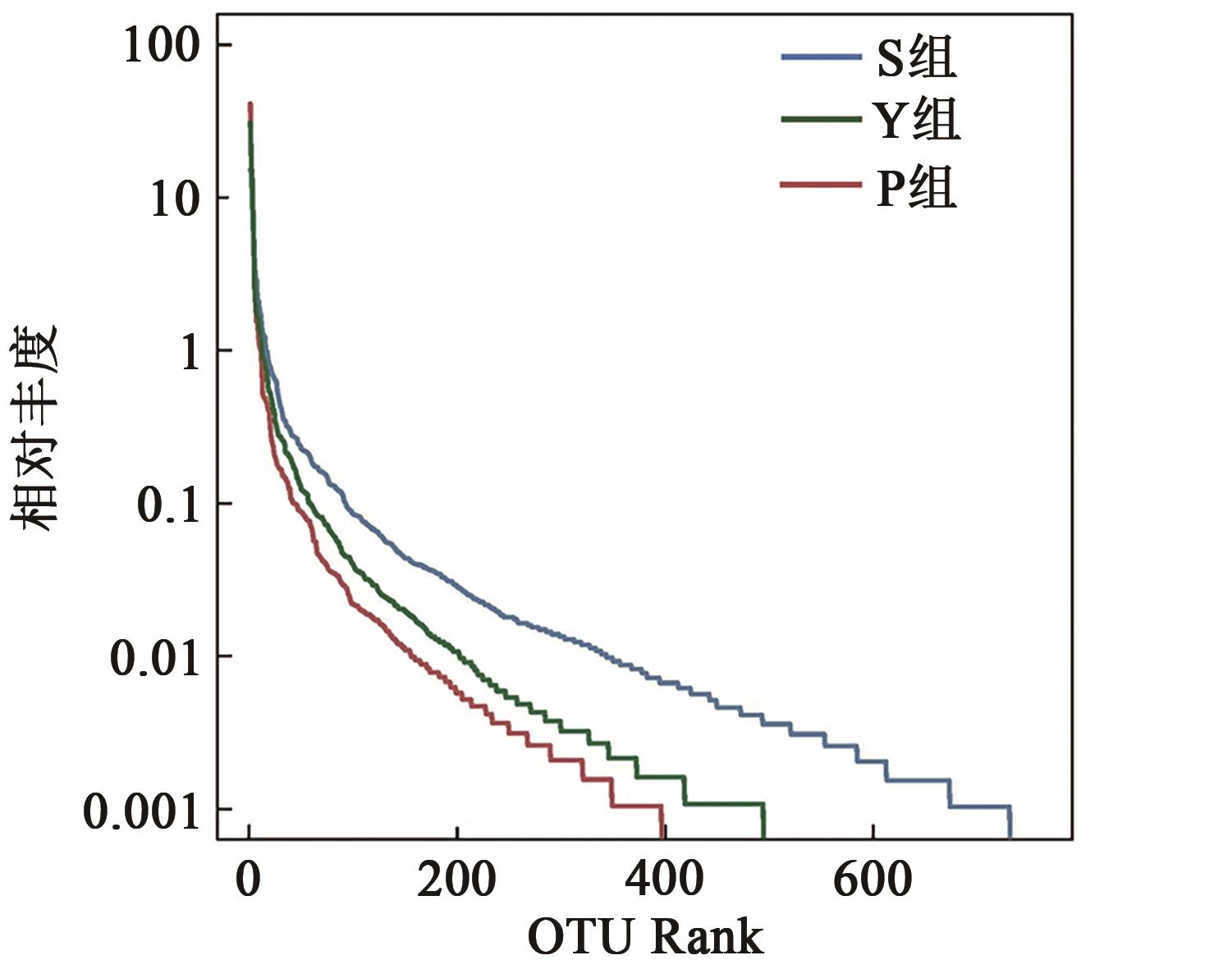
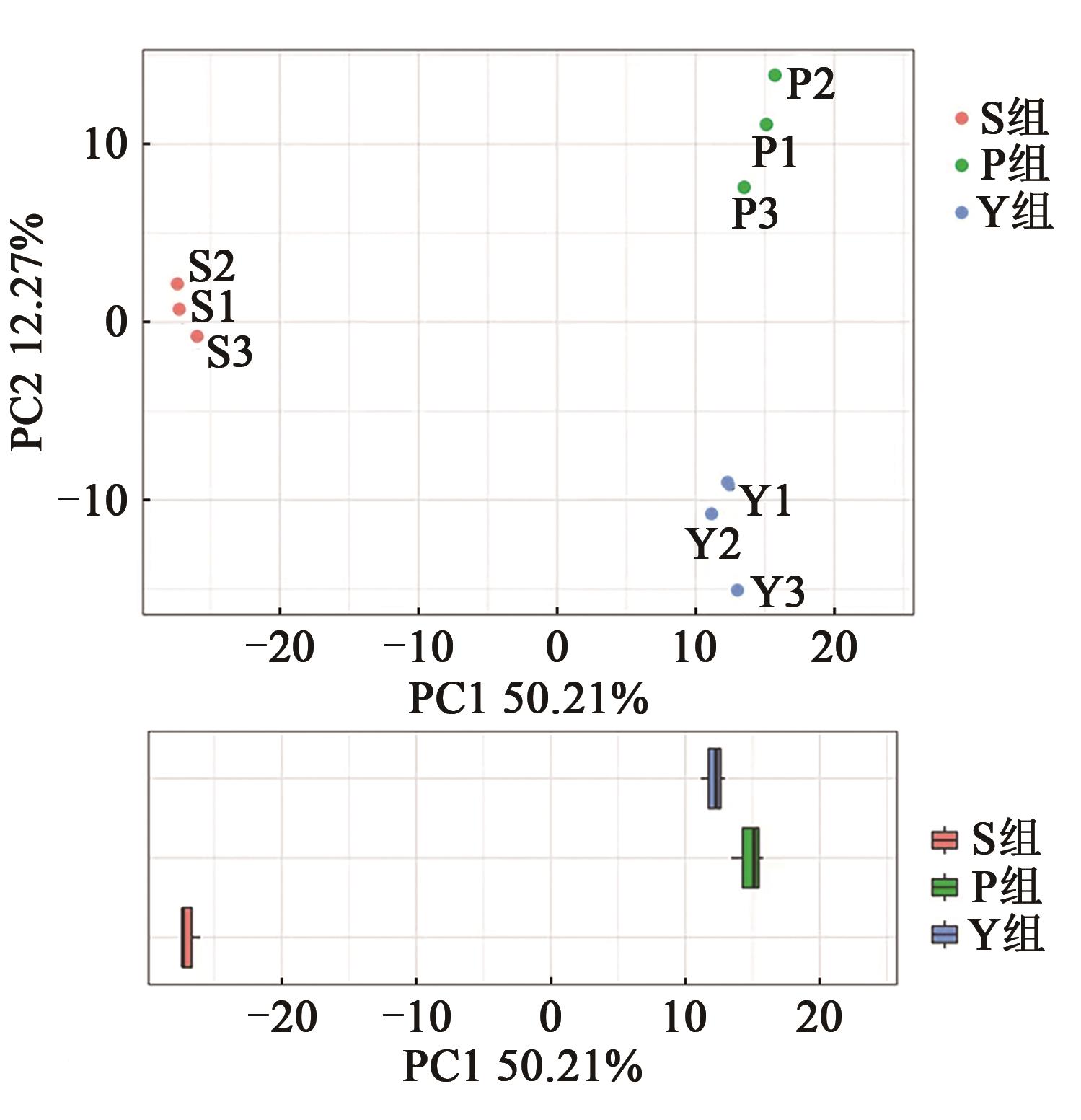
Table 2 Diversity index of faecal flora in different pretreatment groups
| 组别 | Chao 1 | Observed-species | Simpson | Shannon | Goods coverage |
|---|---|---|---|---|---|
| S组 | 707.32 ± 14.090a | 650.17 ± 5.500a | 0.94 ± 0.001a | 5.59 ± 0.030a | 0.998 379a |
| P组 | 418.67 ± 10.430c | 353.07 ± 8.110c | 0.78 ± 0.010c | 3.42 ± 0.050c | 0.998 581a |
| Y组 | 496.76 ± 12.680b | 424.83 ± 3.820b | 0.85 ± 0.020b | 4.12 ± 0.050b | 0.998 225a |
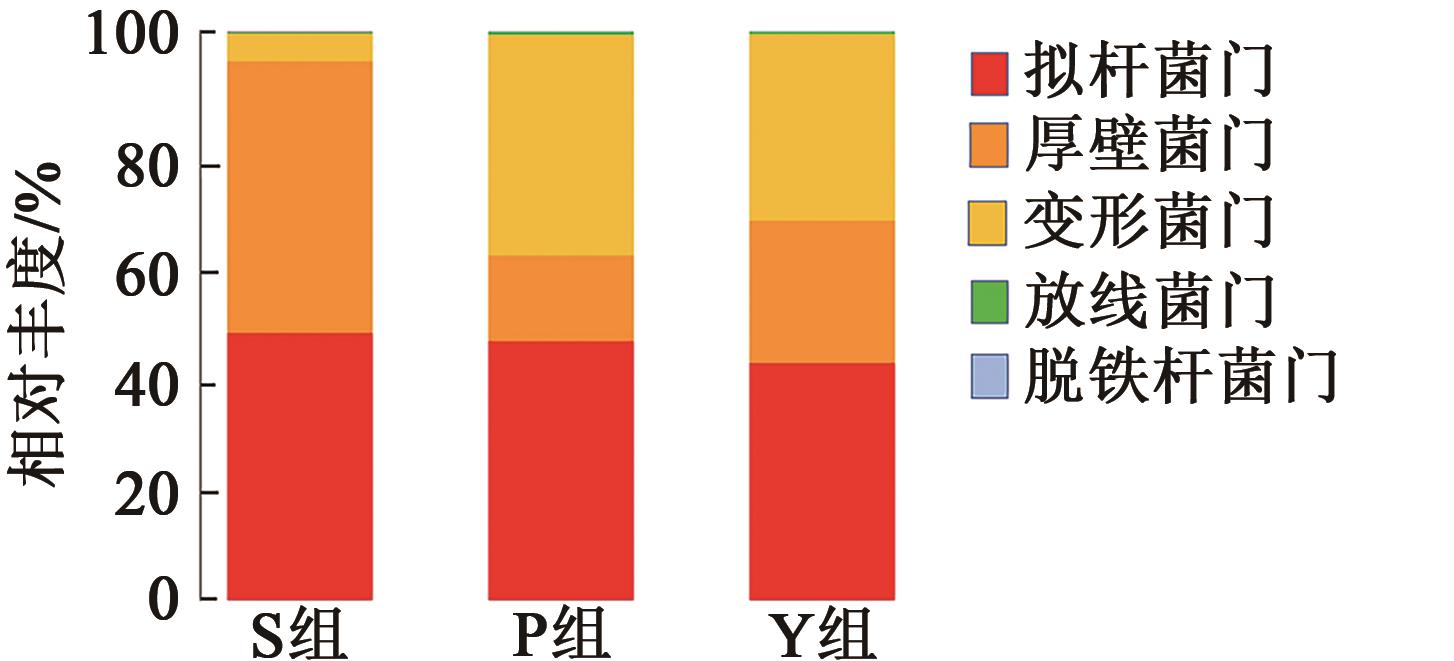
| 组别 | S组 | P组 | Y组 |
|---|---|---|---|
| 拟杆菌门 | 0.469 ± 0.003a | 0.455 ± 0.013a | 0.415 ± 0.056a |
| 厚壁菌门 | 0.478 ± 0.007a | 0.151 ± 0.031c | 0.251 ± 0.035b |
| 变形菌门 | 0.049 ± 0.005b | 0.388 ± 0.046a | 0.330 ± 0.025a |
| 放线菌门 | 0.001 ± 0.000b | 0.004 ± 0.002a | 0.003 ± 0.000ab |
| 脱铁杆菌门 | 0.001 ± 0.000a | 0.001± 0.000a | 0.001 ± 0.000a |
Table 3 Relative abundance of fecal samples in different treatment groups at phylum level(Xˉ±S)
| 组别 | S组 | P组 | Y组 |
|---|---|---|---|
| 拟杆菌门 | 0.469 ± 0.003a | 0.455 ± 0.013a | 0.415 ± 0.056a |
| 厚壁菌门 | 0.478 ± 0.007a | 0.151 ± 0.031c | 0.251 ± 0.035b |
| 变形菌门 | 0.049 ± 0.005b | 0.388 ± 0.046a | 0.330 ± 0.025a |
| 放线菌门 | 0.001 ± 0.000b | 0.004 ± 0.002a | 0.003 ± 0.000ab |
| 脱铁杆菌门 | 0.001 ± 0.000a | 0.001± 0.000a | 0.001 ± 0.000a |
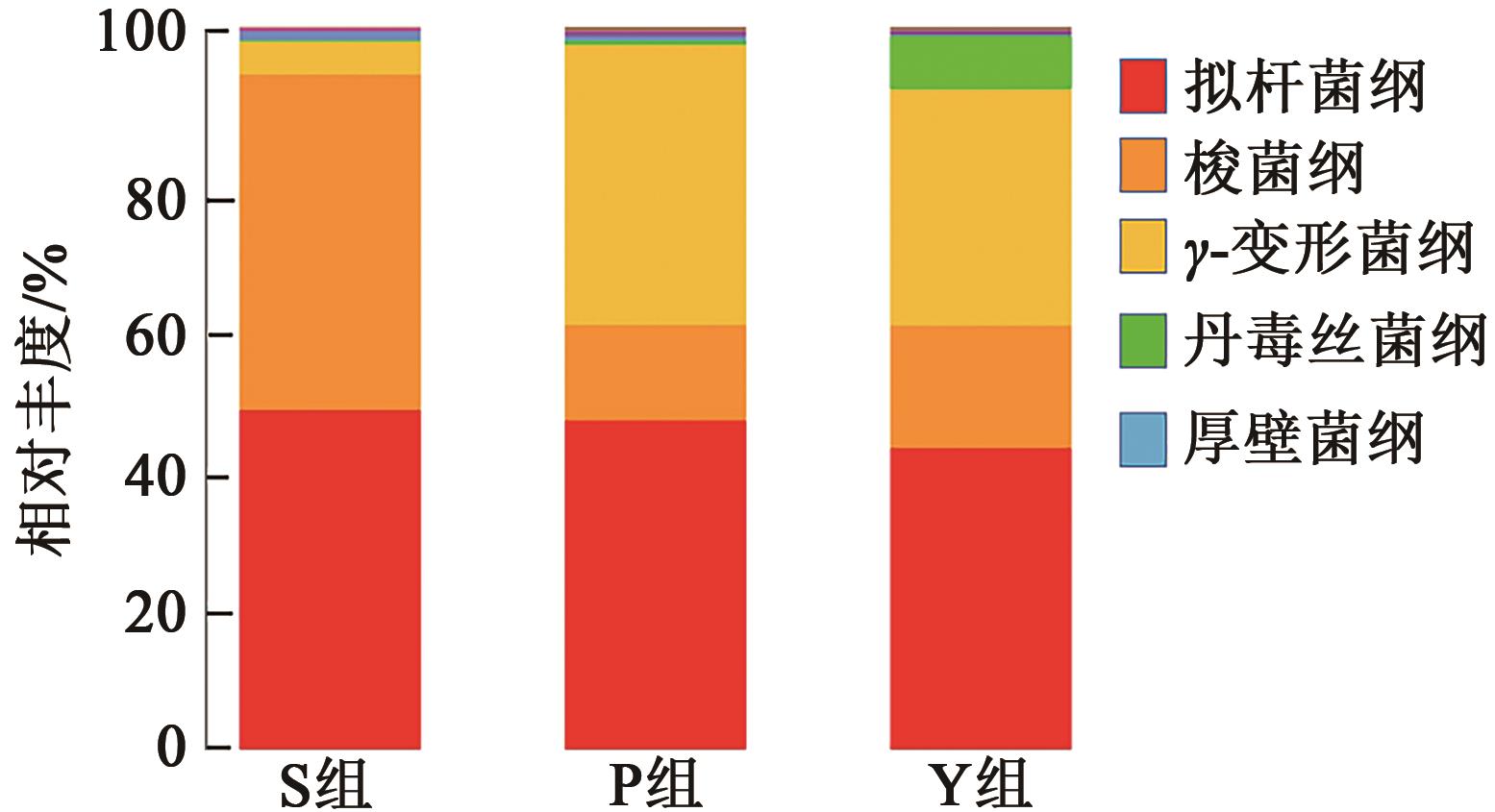
| 组别 | S组 | P组 | Y组 |
|---|---|---|---|
| 拟杆菌纲 | 0.469 ± 0.003a | 0.455 ± 0.013a | 0.415 ± 0.056a |
| 梭菌纲 | 0.464 ± 0.007a | 0.133 ± 0.027b | 0.169 ± 0.016b |
| γ-变形菌纲 | 0.046 ± 0.005a | 0.387 ± 0.046b | 0.329 ± 0.025b |
| 丹毒丝菌纲 | 0.001 ± 0.001b | 0.006 ± 0.001b | 0.074 ± 0.056a |
| 厚壁菌纲 | 0.012 ± 0.000a | 0.007 ± 0.002b | 0.002 ± 0.001c |
Table 4 Relative abundance of fecal samples in different treatment groups at class level(Xˉ±S)
| 组别 | S组 | P组 | Y组 |
|---|---|---|---|
| 拟杆菌纲 | 0.469 ± 0.003a | 0.455 ± 0.013a | 0.415 ± 0.056a |
| 梭菌纲 | 0.464 ± 0.007a | 0.133 ± 0.027b | 0.169 ± 0.016b |
| γ-变形菌纲 | 0.046 ± 0.005a | 0.387 ± 0.046b | 0.329 ± 0.025b |
| 丹毒丝菌纲 | 0.001 ± 0.001b | 0.006 ± 0.001b | 0.074 ± 0.056a |
| 厚壁菌纲 | 0.012 ± 0.000a | 0.007 ± 0.002b | 0.002 ± 0.001c |
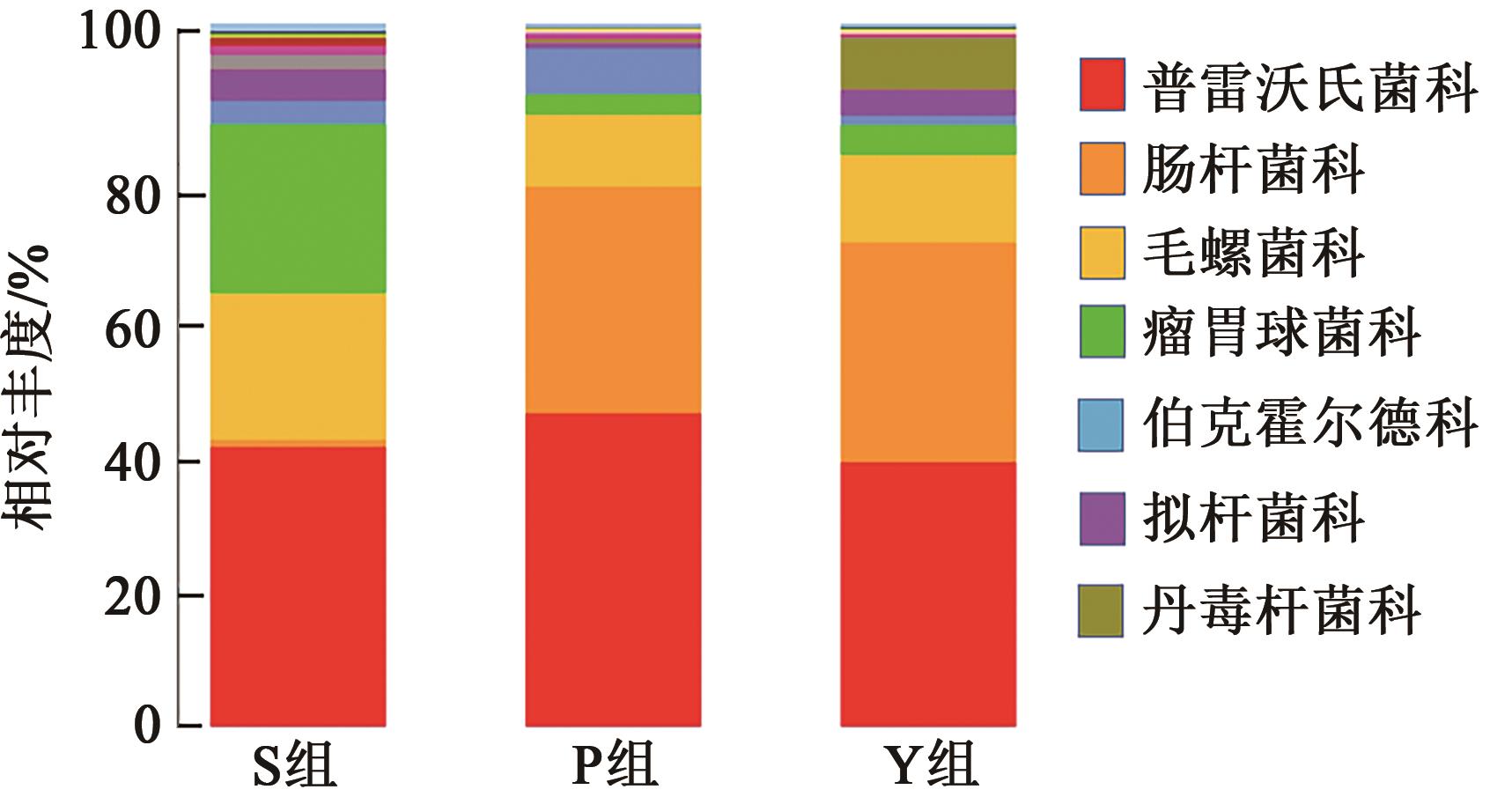
| 组别 | S组 | P组 | Y组 |
|---|---|---|---|
| 普雷沃氏菌科 | 0.397 ± 0.004a | 0.446 ± 0.014a | 0.373 ± 0.061a |
| 肠杆菌科 | 0.010 ± 0.001b | 0.322 ± 0.026a | 0.314 ± 0.038a |
| 毛螺菌科 | 0.210 ± 0.006a | 0.104 ± 0.026b | 0.125 ± 0.02b |
| 瘤胃菌科 | 0.240 ± 0.010a | 0.028 ± 0.002b | 0.042 ± 0.004c |
| 伯克霍尔德科 | 0.034 ± 0.004a | 0.065 ± 0.019b | 0.014 ± 0.014a |
| 拟杆菌科 | 0.046 ± 0.001a | 0.007 ± 0.001c | 0.037 ± 0.008b |
| 丹毒杆菌科 | 0.010 ± 0.000a | 0.006 ± 0.001b | 0.074 ± 0.055a |
Table 5 Relative abundance of fecal samples in different treatment groups at family level(Xˉ±S)
| 组别 | S组 | P组 | Y组 |
|---|---|---|---|
| 普雷沃氏菌科 | 0.397 ± 0.004a | 0.446 ± 0.014a | 0.373 ± 0.061a |
| 肠杆菌科 | 0.010 ± 0.001b | 0.322 ± 0.026a | 0.314 ± 0.038a |
| 毛螺菌科 | 0.210 ± 0.006a | 0.104 ± 0.026b | 0.125 ± 0.02b |
| 瘤胃菌科 | 0.240 ± 0.010a | 0.028 ± 0.002b | 0.042 ± 0.004c |
| 伯克霍尔德科 | 0.034 ± 0.004a | 0.065 ± 0.019b | 0.014 ± 0.014a |
| 拟杆菌科 | 0.046 ± 0.001a | 0.007 ± 0.001c | 0.037 ± 0.008b |
| 丹毒杆菌科 | 0.010 ± 0.000a | 0.006 ± 0.001b | 0.074 ± 0.055a |
| 组别 | S组 | P组 | Y组 |
|---|---|---|---|
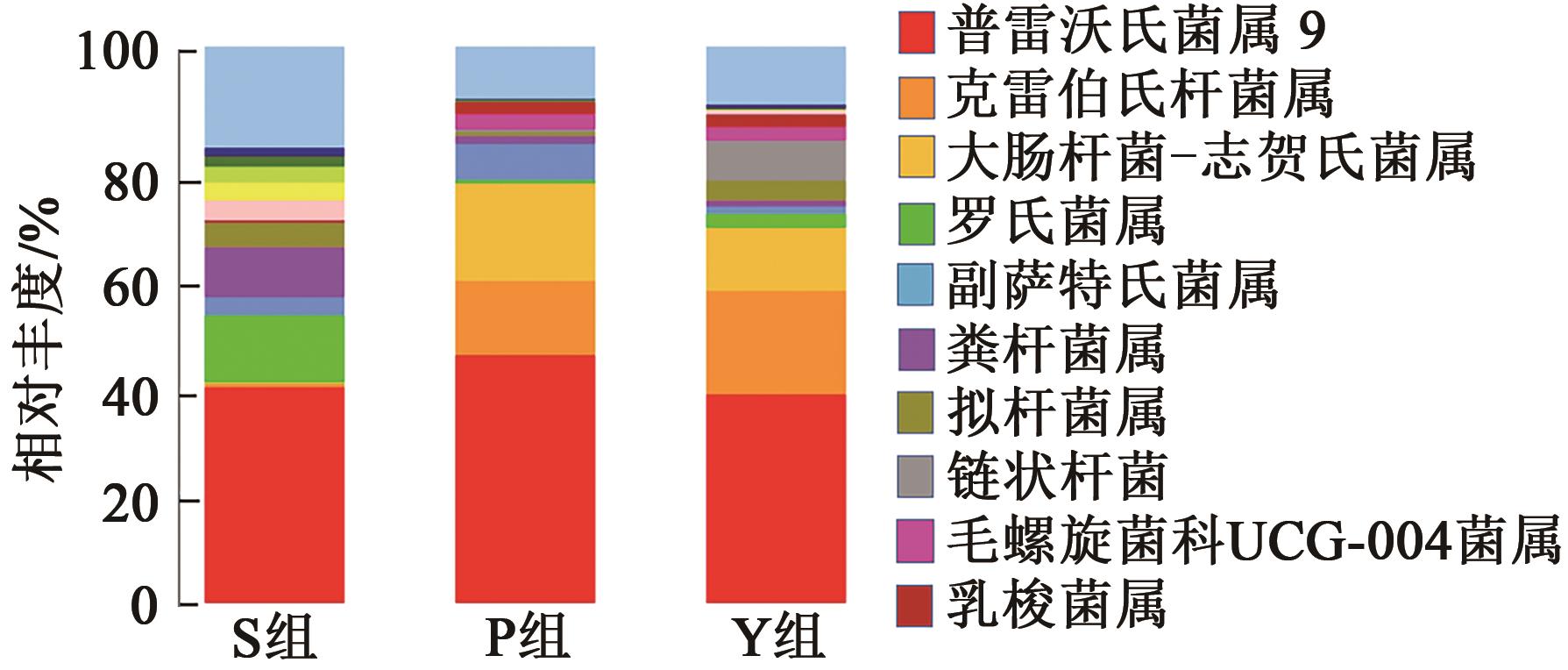
| 普雷沃氏菌属9 | 0.387 ± 0.004a | 0.445 ± 0.014a | 0.372 ± 0.061a |
| 克雷伯氏杆菌属 | 0.007 ± 0.000b | 0.133 ± 0.045a | 0.188 ± 0.038a |
| 大肠杆菌-志贺氏菌属 | 0.003 ± 0.001c | 0.176 ± 0.019a | 0.113 ± 0.008b |
| 罗氏菌属 | 0.120 ± 0.007a | 0.006 ± 0.001b | 0.024 ± 0.018b |
| 粪杆菌属 | 0.089 ± 0.006a | 0.013 ± 0.003b | 0.010 ± 0.004b |
| 副萨特氏菌属 | 0.033 ± 0.005b | 0.065 ± 0.019a | 0.014 ± 0.014b |
| 拟杆菌属 | 0.044 ± 0.002a | 0.007 ± 0.001b | 0.037 ± 0.008a |
| 链状杆菌属 | 0.000 ± 0.000b | 0.005 ± 0.001b | 0.074 ± 0.056a |
| 毛螺旋菌科UCG-004菌属 | 0.000±0.000b | 0.027 ± 0.008a | 0.023 ± 0.004a |
| 乳梭菌属 | 0.003 ± 0.000b | 0.022 ± 0.003a | 0.022 ± 0.003a |
Table 6 Relative abundance of fecal samples in different treatment groups at genus level(Xˉ±S)
| 组别 | S组 | P组 | Y组 |
|---|---|---|---|
| 普雷沃氏菌属9 | 0.387 ± 0.004a | 0.445 ± 0.014a | 0.372 ± 0.061a |
| 克雷伯氏杆菌属 | 0.007 ± 0.000b | 0.133 ± 0.045a | 0.188 ± 0.038a |
| 大肠杆菌-志贺氏菌属 | 0.003 ± 0.001c | 0.176 ± 0.019a | 0.113 ± 0.008b |
| 罗氏菌属 | 0.120 ± 0.007a | 0.006 ± 0.001b | 0.024 ± 0.018b |
| 粪杆菌属 | 0.089 ± 0.006a | 0.013 ± 0.003b | 0.010 ± 0.004b |
| 副萨特氏菌属 | 0.033 ± 0.005b | 0.065 ± 0.019a | 0.014 ± 0.014b |
| 拟杆菌属 | 0.044 ± 0.002a | 0.007 ± 0.001b | 0.037 ± 0.008a |
| 链状杆菌属 | 0.000 ± 0.000b | 0.005 ± 0.001b | 0.074 ± 0.056a |
| 毛螺旋菌科UCG-004菌属 | 0.000±0.000b | 0.027 ± 0.008a | 0.023 ± 0.004a |
| 乳梭菌属 | 0.003 ± 0.000b | 0.022 ± 0.003a | 0.022 ± 0.003a |
| 1 | 王艳丽.紫甘薯酰化花色苷与肠道菌群的相互作用研究[D].天津:天津科技大学,2018. |
| 2 | 王丽凤.益生菌L. plantarum P-8对肉鸡肠道菌群、肠道免疫和生长性能影响的研究[D].呼和浩特:内蒙古农业大学,2014. |
| 3 | 刘嘉奕.人体肠道菌群研究中粪便样品制备与保存研究进展[J].国际儿科学杂志,2019(7):482-485. |
| 4 | 张飞燕,金洁,刘超,等.基于高通量测序技术测定普通棉耳狨猴粪便微生物多样性[J].中国实验动物学报,2019,27(3):366-373. |
| 5 | 董鹏飞.肠道菌群DNA提取与鉴别技术研究[D].天津:天津大学,2016. |
| 6 | 杨成聪,尤利军,吕瑞瑞,等.粪便留取量对粪便中细菌结构和功能的影响[J].中国微生态学杂志,2022,34(6):651-656. |
| 7 | MESSER J W, RICE E W, JOHNSON C H. Spread plate technique[M]//ROBINSON R K, BATT C A, PATEL P D. Encyclopedia of food microbiology. London: Academic Press, 2000: 2159-2166. |
| 8 | GARCIA A, KAMARA J, HAKAN V, et al.. Direct quantification of campylobacter jejuni in chicken fecal samples using real-time PCR: evaluation of six rapid DNA extraction methods[J]. Food Anal. Method., 2013(6): 1728-1738. |
| 9 | 董鹏飞,赵友全,房彦军.最优肠道菌群DNA提取预处理方法的建立[J].中国医药导刊,2016,18(8):849-851. |
| 10 | BABAEI Z, ORMAZDI H, REZAIE S, et al.. Giardia intestinalis: DNA extraction approaches to improve PCR result[J]. Exp. Parasitol., 2011(128): 159-162. |
| 11 | 吴帅帅,武云晖.粪便样本中高纯度人源DNA富集提取方法[J].生物化工,2021,7(6):29-33. |
| 12 | GUPTA N, VERMA V K. Next-generation sequencing and its application: empowering in public health beyond reality[M]// ARORA P. Microbial technology for the welfare of society. Singapore: Springer, 2019: 313-341. |
| 13 | BOLGER A, LOHSE M, USADEL B. Trimmomatic: a flexible trimmer for Illumina sequence data[J]. Bioinformatics, 2014, 30(15): 2114-2120. |
| 14 | MAGOC T, SALZBERG S L. FLASH: fast length adjustment of short reads to improve genome assemblies[J]. Bioinformatics, 2011, 27(21): 2957-2963. |
| 15 | CAPORASO J G, KUCZYNSKI J, STOMBAUGH J, et al.. QIIME allows analysis of high-throughput community sequencing data [J]. Nat. Methods, 2010, 7(5): 335-336. |
| 16 | EDGAR R C, HAAS B J, CLEMENTE J C, et al.. UCHIME improves sensitivity and speed of chimera detection[J]. Bioinformatics, 2011, 27(16): 2194-2200. |
| 17 | 樊英,于晓清,李乐,等.基于16S rRNA高通量测序分析大泷六线鱼表皮粘液及肠道内容物微生物多样性[J].生物技术进展,2021,11(1):79-90. |
| 18 | 刘铭,刘辉,李娜,等.应用高通量测序技术对我国东海带鱼肠道菌群的研究[J].生物技术进展,2020,10(2):152-157. |
| 19 | 申进增.基于16S rDNA测序技术分析当归挥发油对自发性高血压大鼠肠道菌群的影响[D].兰州:甘肃中医药大学,2021. |
| 20 | NEPELSKA M, WOUTERS T, JACOUTON E, et al.. Commensal gut bacteria modulate phosphorylation-dependent PPARγ transcriptional activity in human intestinal epithelial cells[J/OL]. Sci. Rep., 2017, 7: 43199[2022-08-18]. . |
| 21 | 王生,黄晓星,余鹏飞,等.肠道菌群失调与结肠癌发生发展之间关系的研究进展[J].中国药理学通报,2014,30(8):1045-1049. |
| 22 | 张雪雁,李琳琳.一种提取肠道细菌总基因组DNA的方法[J].新疆医科大学学报,2007,30(7):722-724. |
| 23 | 曹冉冉,刘丽,廖虹瑜,等.三种前处理方法对粪便中新型冠状病毒核酸检测结果的影响[J].现代预防医学,2020,47(20):3777-3779. |
| 24 | 黄树武,闵凡贵,王静,等.常见SPF级小鼠和大鼠肠道菌群多样性研究[J].中国实验动物学报,2019,27(2):229-235. |
| 25 | MANICHANH C, B0 RRUEL N, CASELLAS F, et al.. The gut microbiota in IBD[J]. Nat. Rev. Gastroenterol. Hepatol., 2012, 9(10): 599-608. |
| 26 | 赵金香,张瑞十,赵义龙,等.新疆驴盲肠细菌多样性分析[J].草食家畜,2016(4):20-25. |
| 27 | KATSUYAMA Y. Mining novel biosynthetic machineries of secondary metabolites from actinobacteria[J]. Biosci. Biotechnol. Biochem., 2019, 83(9): 1606-1615. |
| 28 | 李琪,孙悦,丁成华,等.泄泻湿证患者肠道菌群特征分析[J].中国中医基础医学杂志,2022,28(1):108-113. |
| 29 | 孙晓霞,周雯,战丽彬.滋补脾阴方药对脾阴虚大鼠肠黏膜屏障的调控作用[J].中国中西医结合杂志,2020,40(10):1233-1240. |
| 30 | ZENG Q, LI D, HE Y, et al.. Discrepant gut microbiota markers for the classification of obesity-related metabolic abnormalities[J]. Sci. Rep., 2019, 9(1): 1-10. |
| 31 | COENYE T. The family burkholderiaceae[M]//ROSENBERG E, DELONG E F, LORY S, et al.. The prokaryotes: alphaproteobacteria and betaproteobacteria. Berlin, Heidelberg: Springer. 2014: 759-776. |
| 32 | 田博雅,马亿珈,奚梦瑶,等.保育期仔猪盲肠微生物菌群多样性分析[J].今日畜牧兽医,2021,37(12):12-15. |
| 33 | QIN J, LI Y, CAI Z, et al.. A metagenome-wide association study of gut microbiota in type 2 diabetes[J]. Nature, 2012, 490(7418): 55-60. |
| 34 | CUI Q L, PAN Y N, XU X T. The metabolic profile of acteoside produced by human or rat intestinal bacteria or intestinal enzyme in vitro employed UPLC-Q-TOF-MS[J]. Fitoterapia, 2016(109): 67-74. |
| 35 | KARLSSON F H, TREMAROLI V, NOOKAEW I, et al.. Gut metagenome in European women with normal, impaired and diabetic glucose control[J]. Nature, 2013, 498(7452): 99-103. |
| [1] | Hui SONG, Wengang CAO, Xiaowen XIAO, Jun DU. Development and Application of Fast NGS Sequencing Method for Plasmid DNA [J]. Current Biotechnology, 2024, 14(4): 594-600. |
| [2] | ZHAO Dongxue1§, LIU Lu2§, MU Yingchun2, HAN Gang2, ZHANG Hongyu2, FANG Hongbo3, RUAN Zhiyong4*, SONG Jinlong2*. Diversity Analysis of Sulfamethoxazole Degradation Community and Isolation and Characterization of Degrading Microorganism [J]. Curr. Biotech., 2021, 11(2): 196-203. |
| [3] | FAN Ying, YU Xiaoqing, LI Le, WANG Xiaolu, YE Haibin, HU Fawen, DIAO Jing*, LIU Hongjun. Analysis of Microbial Diversity of Skin Mucus and Intestinal Contents in Hexagrammos otakii Based on 16S rRNA High-throughput Sequencing Technology [J]. Curr. Biotech., 2021, 11(1): 79-90. |
| [4] | MA Yongkai1, TAO Hongbing1,2, LI Wenru1, XIE Xiaobao1*, SHI Qingshan1, ZHOU Shaolu1. Analysis of Microbial Community Structure and Diversity in Waterborne Coatings [J]. Curr. Biotech., 2019, 9(4): 396-403. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||