Current Biotechnology ›› 2022, Vol. 12 ›› Issue (2): 288-295.DOI: 10.19586/j.2095-2341.2021.0148
• Articles • Previous Articles Next Articles
Analysis of Genetic Polymorphisms at 37 Y⁃STR Loci in Dezhou Han Population
Longfei MIAO1( ), Dapeng SUN1, Wenhua MA2(
), Dapeng SUN1, Wenhua MA2( ), Jianjun ZHONG1(
), Jianjun ZHONG1( )
)
- 1.Material Evidence Authentication and Research Center,Dezhou Public Security Bureau,Shandong Dezhou 253000,China
2.Institute of Forensic Science,Ministry of Public Security,Beijing 100038,China
-
Received:2021-09-03Accepted:2021-11-08Online:2022-03-25Published:2022-03-25 -
Contact:Wenhua MA,Jianjun ZHONG
德州汉族人群37个Y⁃STR基因座遗传多态性分析
- 1.德州市公安局物证鉴定研究中心,山东 德州 253000
2.公安部物证鉴定中心,北京 100038
-
通讯作者:马温华,仲建军 -
作者简介:苗龙飞 E-mail:413695507@qq.com -
基金资助:法医遗传学公安部重点实验室开放课题(2018FGKFKT05)
CLC Number:
Cite this article
Longfei MIAO, Dapeng SUN, Wenhua MA, Jianjun ZHONG. Analysis of Genetic Polymorphisms at 37 Y⁃STR Loci in Dezhou Han Population[J]. Current Biotechnology, 2022, 12(2): 288-295.
苗龙飞, 孙大鹏, 马温华, 仲建军. 德州汉族人群37个Y⁃STR基因座遗传多态性分析[J]. 生物技术进展, 2022, 12(2): 288-295.
share this article
| DYS19 | DYS447 | DYS533 | DYS596 | DYS576 | |||||
|---|---|---|---|---|---|---|---|---|---|
| A | F | A | F | A | F | A | F | A | F |
| 12 | 0.001 7 | 21 | 0.003 3 | 9 | 0.006 7 | 13 | 0.006 7 | 14 | 0.005 0 |
| 13 | 0.055 2 | 22 | 0.015 1 | 10 | 0.063 5 | 14 | 0.361 2 | 15 | 0.006 7 |
| 14 | 0.232 4 | 23 | 0.232 4 | 11 | 0.521 7 | 15 | 0.453 2 | 16 | 0.068 6 |
| 15 | 0.381 3 | 24 | 0.239 1 | 12 | 0.329 4 | 16 | 0.157 2 | 17 | 0.214 0 |
| 15,17 | 0.001 7 | 25 | 0.187 3 | 13 | 0.065 2 | 17 | 0.013 4 | 18 | 0.337 8 |
| 16 | 0.250 8 | 26 | 0.142 1 | 14 | 0.013 4 | 18 | 0.008 4 | 19 | 0.234 1 |
| 17 | 0.071 9 | 27 | 0.123 7 | DYS444 | DYS593 | 20 | 0.080 3 | ||
| 18 | 0.005 0 | 28 | 0.036 8 | A | F | A | F | 21 | 0.045 2 |
| DYS389I | 29 | 0.018 4 | 8 | 0.001 7 | 12 | 0.001 7 | 21,22 | 0.001 7 | |
| A | F | 30 | 0.001 7 | 10 | 0.001 7 | 14 | 0.001 7 | 22 | 0.005 0 |
| 11 | 0.008 4 | DYS449 | 11 | 0.085 3 | 14.2 | 0.001 7 | 24 | 0.001 7 | |
| 12 | 0.500 0 | A | F | 12 | 0.321 1 | 15 | 0.319 4 | DYS438 | |
| 13 | 0.299 3 | 25 | 0.006 7 | 13 | 0.416 4 | 16 | 0.464 9 | A | F |
| 14 | 0.185 6 | 26 | 0.011 7 | 14 | 0.148 8 | 17 | 0.202 3 | 8 | 0.023 4 |
| 15 | 0.006 7 | 27 | 0.045 2 | 15 | 0.023 4 | 18 | 0.008 4 | 9 | 0.011 7 |
| DYS389Ⅱ | 27,28 | 0.001 7 | 16 | 0.0017 | DYS635 | 10 | 0.680 6 | ||
| A | F | 28 | 0.083 6 | DYS557 | A | F | 11 | 0.202 3 | |
| 24 | 0.001 7 | 29 | 0.100 3 | A | F | 18 | 0.003 3 | 12 | 0.066 9 |
| 26 | 0.005 0 | 30 | 0.162 2 | 13 | 0.041 8 | 19 | 0.102 0 | 13 | 0.015 1 |
| 27 | 0.087 0 | 31 | 0.180 6 | 14 | 0.279 3 | 20 | 0.239 1 | DYS439 | |
| 28 | 0.304 3 | 32 | 0.157 2 | 15 | 0.219 1 | 21 | 0.352 8 | A | F |
| 29 | 0.277 6 | 33 | 0.135 5 | 16 | 0.209 0 | 22 | 0.175 6 | 9 | 0.005 0 |
| 30 | 0.225 8 | 33.1 | 0.001 7 | 17 | 0.142 1 | 23 | 0.088 6 | 10 | 0.075 3 |
| 31 | 0.070 2 | 33.2 | 0.001 7 | 17.1 | 0.005 0 | 24 | 0.030 1 | 11 | 0.423 1 |
| 32 | 0.028 4 | 34 | 0.056 9 | 18 | 0.043 5 | 25 | 0.006 7 | 12 | 0.372 9 |
| DYS390 | 35 | 0.046 8 | 19 | 0.045 2 | 26 | 0.001 7 | 13 | 0.108 7 | |
| A | F | 36 | 0.006 7 | 20 | 0.013 4 | DYS643 | 14 | 0.011 7 | |
| 19 | 0.001 7 | 40 | 0.001 7 | 21 | 0.001 7 | A | F | 15 | 0.001 7 |
| 20 | 0.001 7 | DYS456 | DYS570 | 7 | 0.006 7 | 16 | 0.001 7 | ||
| 21 | 0.003 3 | A | F | A | F | 8 | 0.035 1 | DYS385a/b | |
| 22 | 0.035 1 | 13 | 0.025 1 | 14 | 0.018 4 | 9 | 0.098 7 | A | F |
| 23 | 0.431 4 | 14 | 0.180 6 | 15 | 0.028 4 | 10 | 0.314 4 | 12,17 | 0.070 2 |
| 24 | 0.316 1 | 15 | 0.526 8 | 16 | 0.180 6 | 11 | 0.408 0 | 13 | 0.053 5 |
| 25 | 0.192 3 | 16 | 0.205 7 | 17 | 0.224 1 | 12 | 0.115 4 | 12,19 | 0.051 8 |
| 26 | 0.015 1 | 17 | 0.055 2 | 18 | 0.250 8 | 13 | 0.021 7 | 13,19 | 0.040 1 |
| 27 | 0.003 3 | 18 | 0.006 7 | 19 | 0.190 6 | DYS645 | 13,18 | 0.038 5 | |
| DYS391 | DYS458 | 20 | 0.061 9 | A | F | 12,18 | 0.038 5 | ||
| A | F | A | F | 20.3 | 0.003 3 | 7 | 0.003 3 | 11,18 | 0.038 5 |
| 6 | 0.008 4 | 12 | 0.001 7 | 21 | 0.028 4 | 8 | 0.939 8 | 12,16 | 0.038 5 |
| 8 | 0.005 0 | 14 | 0.030 1 | 22 | 0.011 7 | 9 | 0.055 2 | 12,13 | 0.035 1 |
| 9 | 0.065 2 | 15 | 0.100 3 | 23 | 0.001 7 | 10 | 0.001 7 | 12 | 0.035 1 |
| 续表 | |||||||||
| DYS391 | DYS458 | DYF387S1 | GATA_H4 | DYS385a/b | |||||
| A | F | A | F | A | F | A | F | A | F |
| 10 | 0.744 1 | 16 | 0.189 0 | 36,38 | 0.105 4 | 10 | 0.063 5 | 13,20 | 0.031 8 |
| 11 | 0.173 9 | 17 | 0.282 6 | 35,39 | 0.073 6 | 11 | 0.341 1 | 12,15 | 0.030 1 |
| 12 | 0.003 3 | 18 | 0.215 7 | 38 | 0.068 6 | 12 | 0.525 1 | 13,16 | 0.026 8 |
| DYS392 | 19 | 0.107 0 | 35,38 | 0.066 9 | 13 | 0.066 9 | 11,17 | 0.025 1 | |
| A | F | 20 | 0.053 5 | 39 | 0.060 2 | 14 | 0.003 3 | 14,19 | 0.023 4 |
| 7 | 0.001 7 | 21 | 0.011 7 | 37 | 0.055 2 | DYS527a/b | 11,19 | 0.023 4 | |
| 10 | 0.020 1 | 22 | 0.005 0 | 37,38 | 0.048 5 | A | F | 11,16 | 0.023 4 |
| 11 | 0.162 2 | 23 | 0.003 3 | 36,39 | 0.043 5 | 21 | 0.102 0 | 11,12 | 0.023 4 |
| 12 | 0.173 9 | DYS460 | 36,37 | 0.043 5 | 21,23 | 0.095 3 | 11 | 0.021 7 | |
| 13 | 0.329 4 | A | F | 35,37 | 0.043 5 | 20,23 | 0.085 3 | 11,13 | 0.021 7 |
| 14 | 0.269 2 | 8 | 0.005 0 | 37,39 | 0.035 1 | 22 | 0.075 3 | 14,18 | 0.020 1 |
| 15 | 0.038 5 | 9 | 0.275 9 | 36 | 0.031 8 | 21,22 | 0.073 6 | 14,17 | 0.020 1 |
| 16 | 0.005 0 | 10 | 0.408 0 | 36,40 | 0.025 1 | 23 | 0.071 9 | 12,20 | 0.018 4 |
| DYS393 | 11 | 0.280 9 | 38,40 | 0.023 4 | 22,23 | 0.065 2 | 15,17 | 0.016 7 | |
| A | F | 12 | 0.030 1 | 38,39 | 0.023 4 | 20,22 | 0.050 2 | 11,20 | 0.015 1 |
| 8 | 0.001 7 | DYS481 | 35,40 | 0.020 1 | 19,20 | 0.050 2 | 11,14 | 0.013 4 | |
| 11 | 0.005 0 | A | F | 39,40 | 0.020 1 | 20,24 | 0.041 8 | 15,20 | 0.013 4 |
| 12 | 0.526 8 | 19 | 0.003 3 | 35 | 0.018 4 | 21,24 | 0.028 4 | 13,21 | 0.011 7 |
| 12,14 | 0.001 7 | 20 | 0.015 1 | 40 | 0.016 7 | 20 | 0.026 8 | 16,19 | 0.011 7 |
| 13 | 0.239 1 | 21 | 0.041 8 | 37,40 | 0.016 7 | 22,24 | 0.026 8 | 13,17 | 0.010 0 |
| 14 | 0.158 9 | 22 | 0.145 5 | 35,36 | 0.013 4 | 19,21 | 0.025 1 | 13,14 | 0.010 0 |
| 15 | 0.060 2 | 23 | 0.269 2 | 40,41 | 0.013 4 | 19,23 | 0.021 7 | 15,22 | 0.010 0 |
| 16 | 0.006 7 | 24 | 0.239 1 | 36,41 | 0.011 7 | 20,21 | 0.020 1 | 15,23 | 0.008 4 |
| DYS437 | 25 | 0.155 5 | 34 | 0.010 0 | 21,25 | 0.020 1 | 10,19 | 0.008 4 | |
| A | F | 25.1 | 0.001 7 | 34,39 | 0.010 0 | 24 | 0.015 1 | 10,17 | 0.008 4 |
| 11 | 0.003 3 | 26 | 0.068 6 | 35,41 | 0.008 4 | 19,22 | 0.011 7 | 14 | 0.006 7 |
| 13 | 0.010 0 | 27 | 0.028 4 | 34,38 | 0.008 4 | 19 | 0.010 0 | 14,16 | 0.006 7 |
| 14 | 0.607 0 | 28 | 0.015 1 | 37,41 | 0.008 4 | 20,25 | 0.008 4 | 12,14 | 0.006 7 |
| 15 | 0.364 5 | 28.1 | 0.001 7 | 38,41 | 0.008 4 | 23,24 | 0.008 4 | 13,15 | 0.006 7 |
| 16 | 0.015 1 | 29 | 0.010 0 | 34,37 | 0.006 7 | 21,22,23 | 0.006 7 | 16 | 0.006 7 |
| DYS448 | 30 | 0.005 0 | 34,40 | 0.005 0 | 18,24 | 0.006 7 | 15,16 | 0.005 0 | |
| A | F | DYS518 | 41 | 0.005 0 | 18,20 | 0.005 0 | 15,19 | 0.005 0 | |
| 17 | 0.040 1 | A | F | 36,37,38 | 0.005 0 | 22,23,24 | 0.005 0 | 10,12 | 0.005 0 |
| 18 | 0.155 5 | 32 | 0.005 0 | 36,37,40 | 0.003 3 | 21.2,23 | 0.005 0 | 15,21 | 0.005 0 |
| 19 | 0.408 0 | 33 | 0.015 1 | 39,41 | 0.003 3 | 18,26 | 0.005 0 | 12,21 | 0.005 0 |
| 20 | 0.252 5 | 34 | 0.031 8 | 38,42 | 0.003 3 | 19,24 | 0.003 3 | 16,21 | 0.005 0 |
| 21 | 0.118 7 | 35 | 0.113 7 | 35,37,38 | 0.003 3 | 18,21 | 0.003 3 | 14,15 | 0.005 0 |
| 22 | 0.023 4 | 36 | 0.148 8 | 37,39,40 | 0.003 3 | 19,21,23 | 0.003 3 | 10,18 | 0.005 0 |
| 23 | 0.001 7 | 36.2 | 0.015 1 | 38,38 | 0.003 3 | 23,25 | 0.001 7 | 13,22 | 0.003 3 |
| DYS627 | 37 | 0.158 9 | 37,38,39 | 0.003 3 | 20,21,24 | 0.001 7 | 16,20 | 0.003 3 | |
| A | F | 37.2 | 0.028 4 | 34,35,37 | 0.003 3 | 14.3,22 | 0.001 7 | 12,22 | 0.003 3 |
| 16 | 0.006 7 | 38 | 0.152 2 | 37,38,40 | 0.001 7 | 18,19,20 | 0.001 7 | 18,19 | 0.003 3 |
| 续表 | |||||||||
| DYS627 | DYS518 | DYF387S1 | DYS527a/b | DYS385a/b | |||||
| A | F | A | F | A | F | A | F | A | F |
| 17 | 0.038 5 | 38,39 | 0.001 7 | 33 | 0.001 7 | 15.3,23 | 0.001 7 | 14,24 | 0.003 3 |
| 18 | 0.085 3 | 38.2 | 0.005 0 | 36,38,39 | 0.001 7 | 18,23 | 0.001 7 | 11,21 | 0.003 3 |
| 19 | 0.108 7 | 39 | 0.148 8 | 37,40,41 | 0.001 7 | 21,22,23,24 | 0.001 7 | 17,18 | 0.001 7 |
| 19.2 | 0.001 7 | 40 | 0.100 3 | 35,36,39 | 0.001 7 | 33 | 0.001 7 | 13,14,20 | 0.001 7 |
| 20 | 0.183 9 | 41 | 0.046 8 | 36,43 | 0.001 7 | 19,20,22 | 0.001 7 | 10,15 | 0.001 7 |
| 20,21 | 0.001 7 | 42 | 0.023 4 | 37,40,42 | 0.001 7 | 25 | 0.001 7 | 10,21 | 0.001 7 |
| 21 | 0.250 8 | 43 | 0.005 0 | 33,38 | 0.001 7 | 24,26 | 0.001 7 | 12,23 | 0.001 7 |
| 21,22 | 0.001 7 | DYS549 | 36,37,39,40 | 0.001 7 | 23,26 | 0.001 7 | 15,18 | 0.001 7 | |
| 22 | 0.177 3 | A | F | 34,42 | 0.001 7 | 22,25 | 0.001 7 | 11,15 | 0.001 7 |
| 23 | 0.087 0 | 11 | 0.088 6 | 36,40,41 | 0.001 7 | 22,23.1 | 0.001 7 | 13,17,19 | 0.001 7 |
| 24 | 0.050 2 | 12 | 0.533 4 | 34,36 | 0.001 7 | 12,24 | 0.001 7 | ||
| 25 | 0.005 0 | 13 | 0.269 2 | 16,23 | 0.001 7 | ||||
| 26 | 0.001 7 | 14 | 0.090 3 | 17,20 | 0.001 7 | ||||
| 15 | 0.018 4 | 7,12 | 0.001 7 | ||||||
Table 1 Allele frequencies of 37 Y?STR loci in Dezhou Han population (n=598)
| DYS19 | DYS447 | DYS533 | DYS596 | DYS576 | |||||
|---|---|---|---|---|---|---|---|---|---|
| A | F | A | F | A | F | A | F | A | F |
| 12 | 0.001 7 | 21 | 0.003 3 | 9 | 0.006 7 | 13 | 0.006 7 | 14 | 0.005 0 |
| 13 | 0.055 2 | 22 | 0.015 1 | 10 | 0.063 5 | 14 | 0.361 2 | 15 | 0.006 7 |
| 14 | 0.232 4 | 23 | 0.232 4 | 11 | 0.521 7 | 15 | 0.453 2 | 16 | 0.068 6 |
| 15 | 0.381 3 | 24 | 0.239 1 | 12 | 0.329 4 | 16 | 0.157 2 | 17 | 0.214 0 |
| 15,17 | 0.001 7 | 25 | 0.187 3 | 13 | 0.065 2 | 17 | 0.013 4 | 18 | 0.337 8 |
| 16 | 0.250 8 | 26 | 0.142 1 | 14 | 0.013 4 | 18 | 0.008 4 | 19 | 0.234 1 |
| 17 | 0.071 9 | 27 | 0.123 7 | DYS444 | DYS593 | 20 | 0.080 3 | ||
| 18 | 0.005 0 | 28 | 0.036 8 | A | F | A | F | 21 | 0.045 2 |
| DYS389I | 29 | 0.018 4 | 8 | 0.001 7 | 12 | 0.001 7 | 21,22 | 0.001 7 | |
| A | F | 30 | 0.001 7 | 10 | 0.001 7 | 14 | 0.001 7 | 22 | 0.005 0 |
| 11 | 0.008 4 | DYS449 | 11 | 0.085 3 | 14.2 | 0.001 7 | 24 | 0.001 7 | |
| 12 | 0.500 0 | A | F | 12 | 0.321 1 | 15 | 0.319 4 | DYS438 | |
| 13 | 0.299 3 | 25 | 0.006 7 | 13 | 0.416 4 | 16 | 0.464 9 | A | F |
| 14 | 0.185 6 | 26 | 0.011 7 | 14 | 0.148 8 | 17 | 0.202 3 | 8 | 0.023 4 |
| 15 | 0.006 7 | 27 | 0.045 2 | 15 | 0.023 4 | 18 | 0.008 4 | 9 | 0.011 7 |
| DYS389Ⅱ | 27,28 | 0.001 7 | 16 | 0.0017 | DYS635 | 10 | 0.680 6 | ||
| A | F | 28 | 0.083 6 | DYS557 | A | F | 11 | 0.202 3 | |
| 24 | 0.001 7 | 29 | 0.100 3 | A | F | 18 | 0.003 3 | 12 | 0.066 9 |
| 26 | 0.005 0 | 30 | 0.162 2 | 13 | 0.041 8 | 19 | 0.102 0 | 13 | 0.015 1 |
| 27 | 0.087 0 | 31 | 0.180 6 | 14 | 0.279 3 | 20 | 0.239 1 | DYS439 | |
| 28 | 0.304 3 | 32 | 0.157 2 | 15 | 0.219 1 | 21 | 0.352 8 | A | F |
| 29 | 0.277 6 | 33 | 0.135 5 | 16 | 0.209 0 | 22 | 0.175 6 | 9 | 0.005 0 |
| 30 | 0.225 8 | 33.1 | 0.001 7 | 17 | 0.142 1 | 23 | 0.088 6 | 10 | 0.075 3 |
| 31 | 0.070 2 | 33.2 | 0.001 7 | 17.1 | 0.005 0 | 24 | 0.030 1 | 11 | 0.423 1 |
| 32 | 0.028 4 | 34 | 0.056 9 | 18 | 0.043 5 | 25 | 0.006 7 | 12 | 0.372 9 |
| DYS390 | 35 | 0.046 8 | 19 | 0.045 2 | 26 | 0.001 7 | 13 | 0.108 7 | |
| A | F | 36 | 0.006 7 | 20 | 0.013 4 | DYS643 | 14 | 0.011 7 | |
| 19 | 0.001 7 | 40 | 0.001 7 | 21 | 0.001 7 | A | F | 15 | 0.001 7 |
| 20 | 0.001 7 | DYS456 | DYS570 | 7 | 0.006 7 | 16 | 0.001 7 | ||
| 21 | 0.003 3 | A | F | A | F | 8 | 0.035 1 | DYS385a/b | |
| 22 | 0.035 1 | 13 | 0.025 1 | 14 | 0.018 4 | 9 | 0.098 7 | A | F |
| 23 | 0.431 4 | 14 | 0.180 6 | 15 | 0.028 4 | 10 | 0.314 4 | 12,17 | 0.070 2 |
| 24 | 0.316 1 | 15 | 0.526 8 | 16 | 0.180 6 | 11 | 0.408 0 | 13 | 0.053 5 |
| 25 | 0.192 3 | 16 | 0.205 7 | 17 | 0.224 1 | 12 | 0.115 4 | 12,19 | 0.051 8 |
| 26 | 0.015 1 | 17 | 0.055 2 | 18 | 0.250 8 | 13 | 0.021 7 | 13,19 | 0.040 1 |
| 27 | 0.003 3 | 18 | 0.006 7 | 19 | 0.190 6 | DYS645 | 13,18 | 0.038 5 | |
| DYS391 | DYS458 | 20 | 0.061 9 | A | F | 12,18 | 0.038 5 | ||
| A | F | A | F | 20.3 | 0.003 3 | 7 | 0.003 3 | 11,18 | 0.038 5 |
| 6 | 0.008 4 | 12 | 0.001 7 | 21 | 0.028 4 | 8 | 0.939 8 | 12,16 | 0.038 5 |
| 8 | 0.005 0 | 14 | 0.030 1 | 22 | 0.011 7 | 9 | 0.055 2 | 12,13 | 0.035 1 |
| 9 | 0.065 2 | 15 | 0.100 3 | 23 | 0.001 7 | 10 | 0.001 7 | 12 | 0.035 1 |
| 续表 | |||||||||
| DYS391 | DYS458 | DYF387S1 | GATA_H4 | DYS385a/b | |||||
| A | F | A | F | A | F | A | F | A | F |
| 10 | 0.744 1 | 16 | 0.189 0 | 36,38 | 0.105 4 | 10 | 0.063 5 | 13,20 | 0.031 8 |
| 11 | 0.173 9 | 17 | 0.282 6 | 35,39 | 0.073 6 | 11 | 0.341 1 | 12,15 | 0.030 1 |
| 12 | 0.003 3 | 18 | 0.215 7 | 38 | 0.068 6 | 12 | 0.525 1 | 13,16 | 0.026 8 |
| DYS392 | 19 | 0.107 0 | 35,38 | 0.066 9 | 13 | 0.066 9 | 11,17 | 0.025 1 | |
| A | F | 20 | 0.053 5 | 39 | 0.060 2 | 14 | 0.003 3 | 14,19 | 0.023 4 |
| 7 | 0.001 7 | 21 | 0.011 7 | 37 | 0.055 2 | DYS527a/b | 11,19 | 0.023 4 | |
| 10 | 0.020 1 | 22 | 0.005 0 | 37,38 | 0.048 5 | A | F | 11,16 | 0.023 4 |
| 11 | 0.162 2 | 23 | 0.003 3 | 36,39 | 0.043 5 | 21 | 0.102 0 | 11,12 | 0.023 4 |
| 12 | 0.173 9 | DYS460 | 36,37 | 0.043 5 | 21,23 | 0.095 3 | 11 | 0.021 7 | |
| 13 | 0.329 4 | A | F | 35,37 | 0.043 5 | 20,23 | 0.085 3 | 11,13 | 0.021 7 |
| 14 | 0.269 2 | 8 | 0.005 0 | 37,39 | 0.035 1 | 22 | 0.075 3 | 14,18 | 0.020 1 |
| 15 | 0.038 5 | 9 | 0.275 9 | 36 | 0.031 8 | 21,22 | 0.073 6 | 14,17 | 0.020 1 |
| 16 | 0.005 0 | 10 | 0.408 0 | 36,40 | 0.025 1 | 23 | 0.071 9 | 12,20 | 0.018 4 |
| DYS393 | 11 | 0.280 9 | 38,40 | 0.023 4 | 22,23 | 0.065 2 | 15,17 | 0.016 7 | |
| A | F | 12 | 0.030 1 | 38,39 | 0.023 4 | 20,22 | 0.050 2 | 11,20 | 0.015 1 |
| 8 | 0.001 7 | DYS481 | 35,40 | 0.020 1 | 19,20 | 0.050 2 | 11,14 | 0.013 4 | |
| 11 | 0.005 0 | A | F | 39,40 | 0.020 1 | 20,24 | 0.041 8 | 15,20 | 0.013 4 |
| 12 | 0.526 8 | 19 | 0.003 3 | 35 | 0.018 4 | 21,24 | 0.028 4 | 13,21 | 0.011 7 |
| 12,14 | 0.001 7 | 20 | 0.015 1 | 40 | 0.016 7 | 20 | 0.026 8 | 16,19 | 0.011 7 |
| 13 | 0.239 1 | 21 | 0.041 8 | 37,40 | 0.016 7 | 22,24 | 0.026 8 | 13,17 | 0.010 0 |
| 14 | 0.158 9 | 22 | 0.145 5 | 35,36 | 0.013 4 | 19,21 | 0.025 1 | 13,14 | 0.010 0 |
| 15 | 0.060 2 | 23 | 0.269 2 | 40,41 | 0.013 4 | 19,23 | 0.021 7 | 15,22 | 0.010 0 |
| 16 | 0.006 7 | 24 | 0.239 1 | 36,41 | 0.011 7 | 20,21 | 0.020 1 | 15,23 | 0.008 4 |
| DYS437 | 25 | 0.155 5 | 34 | 0.010 0 | 21,25 | 0.020 1 | 10,19 | 0.008 4 | |
| A | F | 25.1 | 0.001 7 | 34,39 | 0.010 0 | 24 | 0.015 1 | 10,17 | 0.008 4 |
| 11 | 0.003 3 | 26 | 0.068 6 | 35,41 | 0.008 4 | 19,22 | 0.011 7 | 14 | 0.006 7 |
| 13 | 0.010 0 | 27 | 0.028 4 | 34,38 | 0.008 4 | 19 | 0.010 0 | 14,16 | 0.006 7 |
| 14 | 0.607 0 | 28 | 0.015 1 | 37,41 | 0.008 4 | 20,25 | 0.008 4 | 12,14 | 0.006 7 |
| 15 | 0.364 5 | 28.1 | 0.001 7 | 38,41 | 0.008 4 | 23,24 | 0.008 4 | 13,15 | 0.006 7 |
| 16 | 0.015 1 | 29 | 0.010 0 | 34,37 | 0.006 7 | 21,22,23 | 0.006 7 | 16 | 0.006 7 |
| DYS448 | 30 | 0.005 0 | 34,40 | 0.005 0 | 18,24 | 0.006 7 | 15,16 | 0.005 0 | |
| A | F | DYS518 | 41 | 0.005 0 | 18,20 | 0.005 0 | 15,19 | 0.005 0 | |
| 17 | 0.040 1 | A | F | 36,37,38 | 0.005 0 | 22,23,24 | 0.005 0 | 10,12 | 0.005 0 |
| 18 | 0.155 5 | 32 | 0.005 0 | 36,37,40 | 0.003 3 | 21.2,23 | 0.005 0 | 15,21 | 0.005 0 |
| 19 | 0.408 0 | 33 | 0.015 1 | 39,41 | 0.003 3 | 18,26 | 0.005 0 | 12,21 | 0.005 0 |
| 20 | 0.252 5 | 34 | 0.031 8 | 38,42 | 0.003 3 | 19,24 | 0.003 3 | 16,21 | 0.005 0 |
| 21 | 0.118 7 | 35 | 0.113 7 | 35,37,38 | 0.003 3 | 18,21 | 0.003 3 | 14,15 | 0.005 0 |
| 22 | 0.023 4 | 36 | 0.148 8 | 37,39,40 | 0.003 3 | 19,21,23 | 0.003 3 | 10,18 | 0.005 0 |
| 23 | 0.001 7 | 36.2 | 0.015 1 | 38,38 | 0.003 3 | 23,25 | 0.001 7 | 13,22 | 0.003 3 |
| DYS627 | 37 | 0.158 9 | 37,38,39 | 0.003 3 | 20,21,24 | 0.001 7 | 16,20 | 0.003 3 | |
| A | F | 37.2 | 0.028 4 | 34,35,37 | 0.003 3 | 14.3,22 | 0.001 7 | 12,22 | 0.003 3 |
| 16 | 0.006 7 | 38 | 0.152 2 | 37,38,40 | 0.001 7 | 18,19,20 | 0.001 7 | 18,19 | 0.003 3 |
| 续表 | |||||||||
| DYS627 | DYS518 | DYF387S1 | DYS527a/b | DYS385a/b | |||||
| A | F | A | F | A | F | A | F | A | F |
| 17 | 0.038 5 | 38,39 | 0.001 7 | 33 | 0.001 7 | 15.3,23 | 0.001 7 | 14,24 | 0.003 3 |
| 18 | 0.085 3 | 38.2 | 0.005 0 | 36,38,39 | 0.001 7 | 18,23 | 0.001 7 | 11,21 | 0.003 3 |
| 19 | 0.108 7 | 39 | 0.148 8 | 37,40,41 | 0.001 7 | 21,22,23,24 | 0.001 7 | 17,18 | 0.001 7 |
| 19.2 | 0.001 7 | 40 | 0.100 3 | 35,36,39 | 0.001 7 | 33 | 0.001 7 | 13,14,20 | 0.001 7 |
| 20 | 0.183 9 | 41 | 0.046 8 | 36,43 | 0.001 7 | 19,20,22 | 0.001 7 | 10,15 | 0.001 7 |
| 20,21 | 0.001 7 | 42 | 0.023 4 | 37,40,42 | 0.001 7 | 25 | 0.001 7 | 10,21 | 0.001 7 |
| 21 | 0.250 8 | 43 | 0.005 0 | 33,38 | 0.001 7 | 24,26 | 0.001 7 | 12,23 | 0.001 7 |
| 21,22 | 0.001 7 | DYS549 | 36,37,39,40 | 0.001 7 | 23,26 | 0.001 7 | 15,18 | 0.001 7 | |
| 22 | 0.177 3 | A | F | 34,42 | 0.001 7 | 22,25 | 0.001 7 | 11,15 | 0.001 7 |
| 23 | 0.087 0 | 11 | 0.088 6 | 36,40,41 | 0.001 7 | 22,23.1 | 0.001 7 | 13,17,19 | 0.001 7 |
| 24 | 0.050 2 | 12 | 0.533 4 | 34,36 | 0.001 7 | 12,24 | 0.001 7 | ||
| 25 | 0.005 0 | 13 | 0.269 2 | 16,23 | 0.001 7 | ||||
| 26 | 0.001 7 | 14 | 0.090 3 | 17,20 | 0.001 7 | ||||
| 15 | 0.018 4 | 7,12 | 0.001 7 | ||||||
| 基因座 | GD值 | 基因座 | GD值 | 基因座 | GD值 | 基因座 | GD值 | 基因座 | GD值 |
|---|---|---|---|---|---|---|---|---|---|
| DYS385a/b | 0.971 4 | DYS447 | 0.817 6 | DYS392 | 0.761 8 | DYS439 | 0.665 4 | DYS533 | 0.611 8 |
| DYF387S1 | 0.953 8 | DYS458 | 0.813 8 | DYS19 | 0.730 7 | DYS456 | 0.645 0 | GATA_H4 | 0.600 4 |
| DYS527a/b | 0.942 3 | DYS570 | 0.813 3 | DYS448 | 0.730 5 | DYS593 | 0.641 9 | DYS437 | 0.499 1 |
| DYS518 | 0.880 7 | DYS557 | 0.805 6 | DYS643 | 0.711 1 | DYS596 | 0.640 2 | DYS438 | 0.491 3 |
| DYS449 | 0.874 7 | DYS576 | 0.773 3 | DYS444 | 0.694 7 | DYS393 | 0.637 5 | DYS391 | 0.412 3 |
| DYS627 | 0.842 5 | DYS635 | 0.769 6 | DYS460 | 0.678 7 | DYS549 | 0.627 7 | DYS645 | 0.113 9 |
| DYS481 | 0.818 5 | DYS38Ⅱ | 0.767 3 | DYS390 | 0.676 6 | DYS389I | 0.626 9 |
Table 2 Gene diversity of Dezhou Han population(n=598)
| 基因座 | GD值 | 基因座 | GD值 | 基因座 | GD值 | 基因座 | GD值 | 基因座 | GD值 |
|---|---|---|---|---|---|---|---|---|---|
| DYS385a/b | 0.971 4 | DYS447 | 0.817 6 | DYS392 | 0.761 8 | DYS439 | 0.665 4 | DYS533 | 0.611 8 |
| DYF387S1 | 0.953 8 | DYS458 | 0.813 8 | DYS19 | 0.730 7 | DYS456 | 0.645 0 | GATA_H4 | 0.600 4 |
| DYS527a/b | 0.942 3 | DYS570 | 0.813 3 | DYS448 | 0.730 5 | DYS593 | 0.641 9 | DYS437 | 0.499 1 |
| DYS518 | 0.880 7 | DYS557 | 0.805 6 | DYS643 | 0.711 1 | DYS596 | 0.640 2 | DYS438 | 0.491 3 |
| DYS449 | 0.874 7 | DYS576 | 0.773 3 | DYS444 | 0.694 7 | DYS393 | 0.637 5 | DYS391 | 0.412 3 |
| DYS627 | 0.842 5 | DYS635 | 0.769 6 | DYS460 | 0.678 7 | DYS549 | 0.627 7 | DYS645 | 0.113 9 |
| DYS481 | 0.818 5 | DYS38Ⅱ | 0.767 3 | DYS390 | 0.676 6 | DYS389I | 0.626 9 |
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | + | + | + | + | + | - | + | + | + | + | + | + | + | + | - | |
| 2 | 0.103 1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| 3 | 0.063 7 | 0.055 1 | + | + | + | + | + | + | + | + | + | + | - | + | + | |
| 4 | 0.016 1 | 0.070 2 | 0.056 3 | + | + | + | + | + | + | + | + | + | + | + | + | |
| 5 | 0.014 9 | 0.152 9 | 0.100 2 | 0.028 2 | + | + | + | + | + | + | + | + | + | + | - | |
| 6 | 0.006 0 | 0.111 0 | 0.071 8 | 0.012 6 | 0.006 9 | + | + | + | + | + | + | + | + | + | - | |
| 7 | 0.012 1 | 0.107 0 | 0.075 9 | 0.011 6 | 0.015 9 | 0.002 8 | + | + | + | + | + | + | + | + | - | |
| 8 | 0.004 9 | 0.108 7 | 0.065 0 | 0.015 1 | 0.010 5 | 0.011 4 | 0.023 4 | - | + | + | + | + | + | + | - | |
| 9 | 0.002 9 | 0.099 2 | 0.066 5 | 0.016 3 | 0.012 0 | 0.008 3 | 0.015 4 | 0.003 7 | + | + | + | + | + | + | - | |
| 10 | 0.032 2 | 0.066 7 | 0.007 1 | 0.030 1 | 0.060 5 | 0.039 9 | 0.045 7 | 0.030 5 | 0.036 0 | + | + | + | + | + | + | |
| 11 | 0.075 7 | 0.092 6 | 0.092 1 | 0.029 6 | 0.087 5 | 0.049 7 | 0.036 4 | 0.087 8 | 0.076 2 | 0.072 3 | + | + | + | + | + | |
| 12 | 0.040 1 | 0.029 3 | 0.031 3 | 0.031 2 | 0.073 0 | 0.053 3 | 0.060 3 | 0.035 9 | 0.039 2 | 0.025 2 | 0.088 9 | + | + | + | + | |
| 13 | 0.052 3 | 0.119 2 | 0.087 1 | 0.049 3 | 0.072 8 | 0.051 0 | 0.046 4 | 0.071 4 | 0.063 0 | 0.064 0 | 0.072 3 | 0.075 3 | + | + | + | |
| 14 | 0.067 2 | 0.080 5 | 0.003 3 | 0.068 7 | 0.107 7 | 0.078 0 | 0.085 3 | 0.063 6 | 0.068 0 | 0.006 1 | 0.117 2 | 0.044 6 | 0.103 7 | + | + | |
| 15 | 0.011 5 | 0.105 3 | 0.075 5 | 0.010 0 | 0.010 6 | 0.001 6 | 0.002 0 | 0.018 5 | 0.011 9 | 0.045 3 | 0.044 6 | 0.053 4 | 0.054 7 | 0.085 7 | - | |
| 16 | 0.009 1 | 0.166 4 | 0.104 6 | 0.042 5 | 0.008 1 | 0.008 1 | 0.014 1 | 0.015 4 | 0.010 9 | 0.064 4 | 0.106 5 | 0.090 8 | 0.073 6 | 0.101 1 | 0.016 0 |
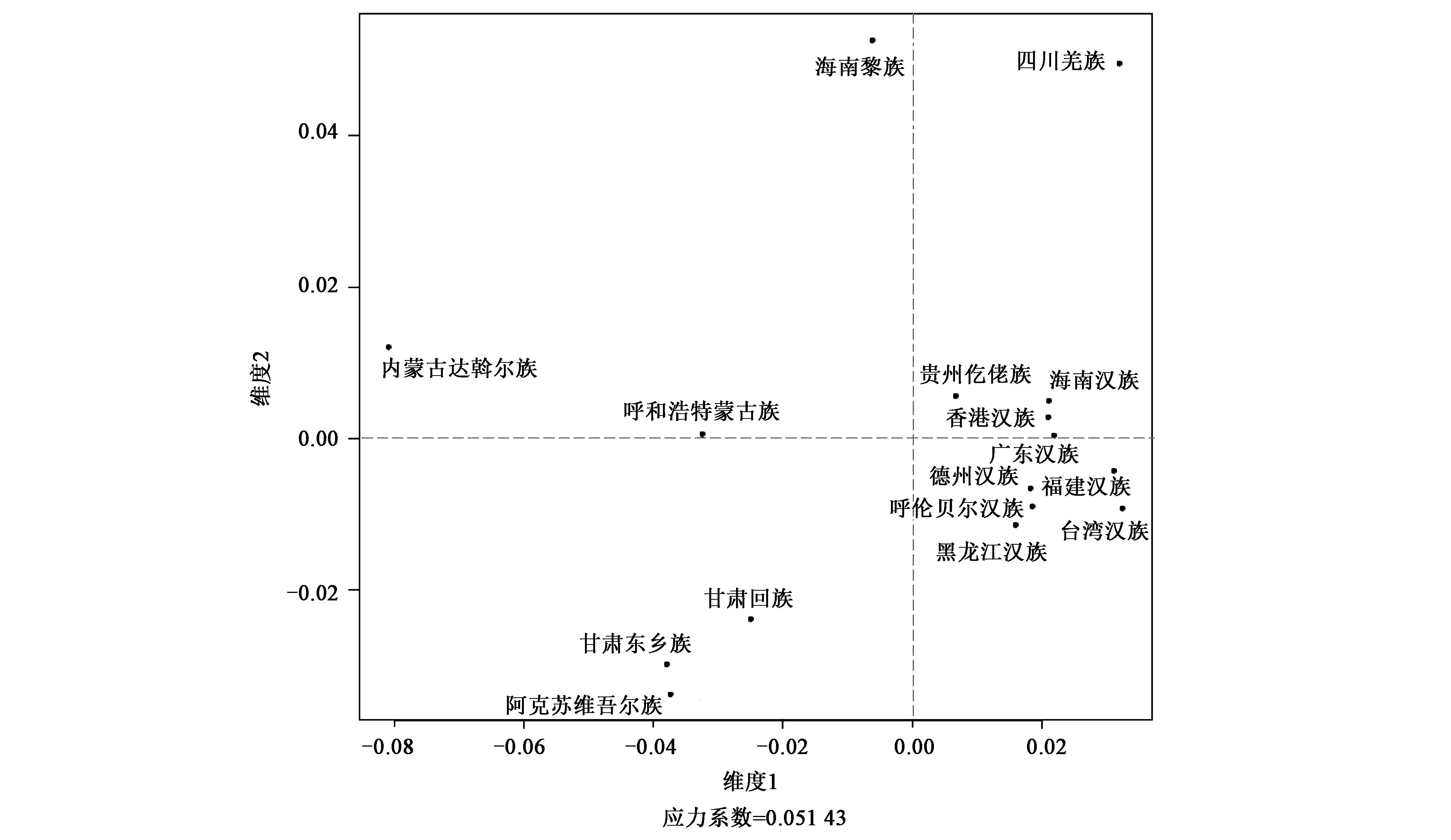
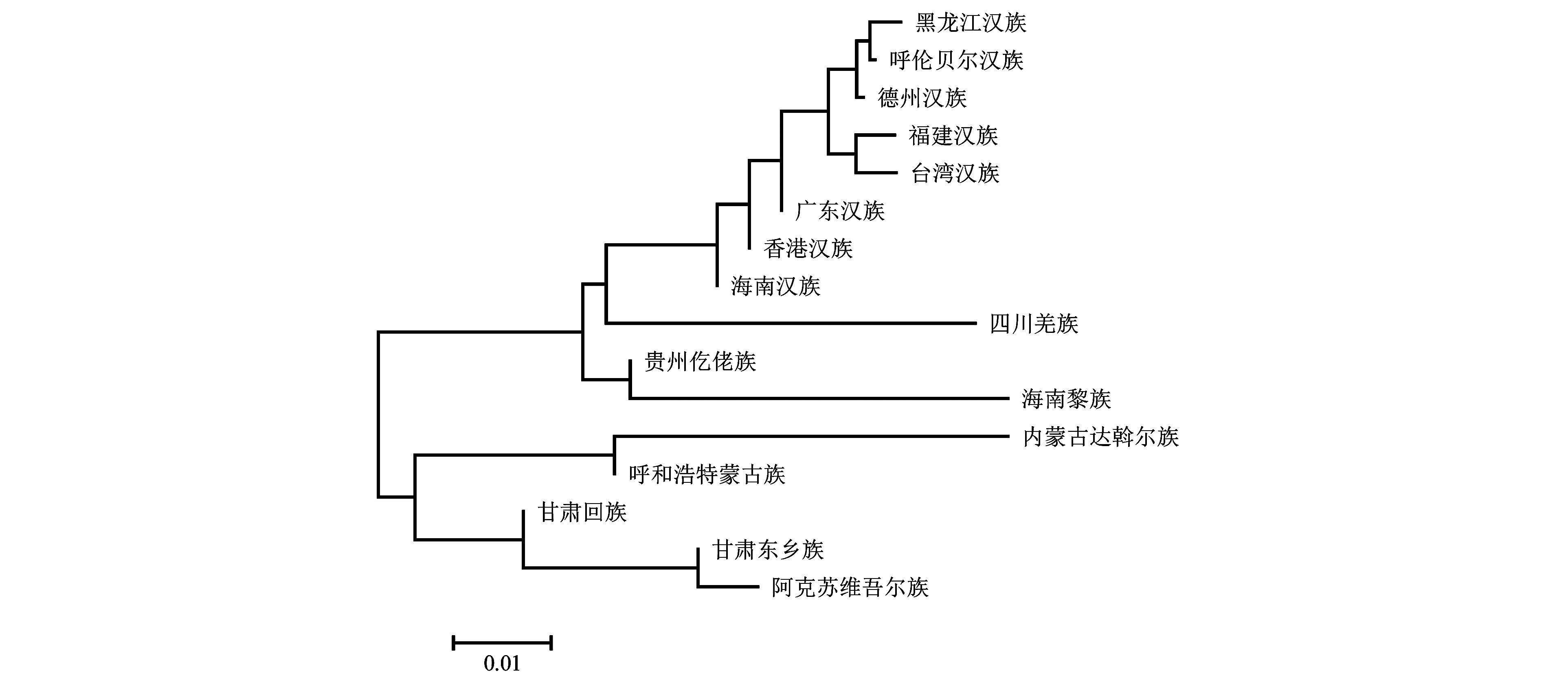
Table 3 Genetic distance matrix of Rst values between 16 populations
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | + | + | + | + | + | - | + | + | + | + | + | + | + | + | - | |
| 2 | 0.103 1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| 3 | 0.063 7 | 0.055 1 | + | + | + | + | + | + | + | + | + | + | - | + | + | |
| 4 | 0.016 1 | 0.070 2 | 0.056 3 | + | + | + | + | + | + | + | + | + | + | + | + | |
| 5 | 0.014 9 | 0.152 9 | 0.100 2 | 0.028 2 | + | + | + | + | + | + | + | + | + | + | - | |
| 6 | 0.006 0 | 0.111 0 | 0.071 8 | 0.012 6 | 0.006 9 | + | + | + | + | + | + | + | + | + | - | |
| 7 | 0.012 1 | 0.107 0 | 0.075 9 | 0.011 6 | 0.015 9 | 0.002 8 | + | + | + | + | + | + | + | + | - | |
| 8 | 0.004 9 | 0.108 7 | 0.065 0 | 0.015 1 | 0.010 5 | 0.011 4 | 0.023 4 | - | + | + | + | + | + | + | - | |
| 9 | 0.002 9 | 0.099 2 | 0.066 5 | 0.016 3 | 0.012 0 | 0.008 3 | 0.015 4 | 0.003 7 | + | + | + | + | + | + | - | |
| 10 | 0.032 2 | 0.066 7 | 0.007 1 | 0.030 1 | 0.060 5 | 0.039 9 | 0.045 7 | 0.030 5 | 0.036 0 | + | + | + | + | + | + | |
| 11 | 0.075 7 | 0.092 6 | 0.092 1 | 0.029 6 | 0.087 5 | 0.049 7 | 0.036 4 | 0.087 8 | 0.076 2 | 0.072 3 | + | + | + | + | + | |
| 12 | 0.040 1 | 0.029 3 | 0.031 3 | 0.031 2 | 0.073 0 | 0.053 3 | 0.060 3 | 0.035 9 | 0.039 2 | 0.025 2 | 0.088 9 | + | + | + | + | |
| 13 | 0.052 3 | 0.119 2 | 0.087 1 | 0.049 3 | 0.072 8 | 0.051 0 | 0.046 4 | 0.071 4 | 0.063 0 | 0.064 0 | 0.072 3 | 0.075 3 | + | + | + | |
| 14 | 0.067 2 | 0.080 5 | 0.003 3 | 0.068 7 | 0.107 7 | 0.078 0 | 0.085 3 | 0.063 6 | 0.068 0 | 0.006 1 | 0.117 2 | 0.044 6 | 0.103 7 | + | + | |
| 15 | 0.011 5 | 0.105 3 | 0.075 5 | 0.010 0 | 0.010 6 | 0.001 6 | 0.002 0 | 0.018 5 | 0.011 9 | 0.045 3 | 0.044 6 | 0.053 4 | 0.054 7 | 0.085 7 | - | |
| 16 | 0.009 1 | 0.166 4 | 0.104 6 | 0.042 5 | 0.008 1 | 0.008 1 | 0.014 1 | 0.015 4 | 0.010 9 | 0.064 4 | 0.106 5 | 0.090 8 | 0.073 6 | 0.101 1 | 0.016 0 |
| 1 | 郑秀芬. 法医DNA分析[M]. 北京:中国人民公安大学出版社, 2002: 300-301. |
| 2 | KAYSER M. The Human Y-chromosome-introduction into genetics and applications [J]. Forensic Sci. Rev., 2003, 15(2): 77-89. |
| 3 | ROEWER L, NURNBERG P, FUHRMANN E, et al.. Stain analysis using oligonucleotide probes specific for simple repetitive DNA sequences [J]. Forensic Sci. Int., 1990, 47(1): 59-70. |
| 4 | KAYSER M, BRAUER S, WEISS G, et al.. Independent histories of human Y chromosomes from Melanesia and Australia[J]. Am. J. Hum. Genet., 2001, 68(1): 173-190. |
| 5 | KAYSER M, BRAUER S, WEISS G, et al.. Melanesian origin of polynesian Y chromosomes [J]. Curr. Biol., 2000, 10(20): 1237-1246. |
| 6 | JOBLING M A, TYLER-SMITH C. Father and sons: the Y chromosome and human evolution[J]. Trends Genet., 1995,11:449-456. |
| 7 | 师文远, 王瑛, 李海霞, 等. 基于Y-STR分型的中国汉族群体遗传关系[J]. 热带医学杂志, 2016,16(3):281-284. |
| 8 | 邓盼, 江丽, 马泉, 等. 广西6个民族24个Y-STR基因座遗传多态性及群体遗传结构分析[J]. 刑事技术, 2018,43(3 ):198-201. |
| 9 | 尚蕾, 莫晓婷, 张建, 等. 周口地区汉族人群26个Y-STR基因座的遗传多态性[J]. 激光生物学报, 2018, 27(1): 91-96. |
| 10 | 孟娟, 王晓丹, 黄艳梅, 等. 河南新乡地区汉族人群27个Y-STR基因座遗传多态性检测[J].郑州大学学报, 2018, 53(4): 452-457. |
| 11 | 赵霖, 李晓明, 付永, 等. 湖南长沙汉族人群25个Y-STR基因座的遗传多态性[J]. 法医学杂志, 2018, 34(1): 79-81. |
| 12 | 刘亚举, 孙现锋, 岳俊涛, 等. 中国汉族群体36个Y-STR基因座的遗传多态性及突变研究[J].中国输血杂志, 2018, 31(5):495-499. |
| 13 | ZEYE M M J, LI J, OUEDRAOGO S Y, et al.. Population data and genetic structure analysis based on 29 Y-STR loci among the ethnolinguistic groups in Burkina Faso[J]. Int. J. Legal Med., 2021,135(5): 1767-1769. |
| 14 | KUMAR S, STECHER G, TAMURA K. MEGA7: molecular evolutionary genetics analysis Version 7.0 for bigger datasets[J]. Mol. Biol. Evol., 2016,33(7) : 1870-1874. |
| 15 | SAITOU N,NEI M. The neighbor-joining method: a new method for reconstructing phylogenetic trees[J]. Mol.Biol.Evol.,1987, 4: 406-425. |
| 16 | ALGHAFRI R, ZUPANIC P I, ZUPANC T, et al.. Rapidly mutating Y-STR analyses of compromised forensic samples [J]. Int. J. Legal. Med., 2018, 132(2): 397-403. |
| 17 | BALLANTYNE K N, KEEL V, WOLLSTEIN A, et al.. A new future of forensic Y-chromosome analysis: rapidly mutating Y-STRs for differentiating male relatives and paternal lineages[J]. Forensic Sci. Int. Genet.,2012,6(2):208-218. |
| 18 | 吴微微, 张晓霞, 金雷, 等. 中国汉族男性家系Y-STR 基因座容差情况调查[J].中国法医学杂志, 2020,35(4):390-393. |
| 19 | ZHANG F, SU B, ZHANG Y, et al.. Genetic studies of human diversity in East Asia [J]. Phil. Trans. R. Soc. B, 2007, 362(1482): 987-996. |
| 20 | 蔡晓云.Y染色体揭示的早期人类进入东亚和东亚人群特征形成过程[D].上海:复旦大学, 2009. |
| 21 | ROSENBERG N A, PRITECHARD J K, WEBER J L, et al.. Genetic structure of human populations[J]. Science, 2002,298(5602):2381-2385. |
| 22 | ATKINSON Q D. Phonemic diversity supports a serial founder effect model of language expansion from Africa[J]. Science, 2011, 332(6027): 346-349. |
| 23 | 许正文.论历史时期大陆向台湾的移民与往来[J].中国历史地理论丛, 2001(16)3:67-71. |
| 24 | 张绚光. 二十世纪后半期回族史学论述[D]. 兰州: 兰州大学, 2012. |
| 25 | ROSENBERG N A, PRITCHARD J K, WEBER J L, et a1.. Genetics structure of human populations[J]. Science, 2002, 298(5602): 2381-2385. |
| 26 | ROSENBERG N A, MAHAJAN S, RAMACHANDRAN S,et a1.. Clines,clusters,and the effect of study design on the inference of human populations structure[J]. PLoS Genet., 2005, 1(6): 660-671. |
| 27 | SHRIVER M D, MEI R, PARRA E J,et a1..Largescale SNP analysis reveals clustered and continuous patterns of human genetic variation[J]. Hum. Genom., 2005, 2(2):81-89. |
| 28 | KAYSER M, KRAWCZAK M, EXCOFFIER L, et al.. An extensive analysis of Y-chromosomal microsatellite haplotypes in globally dispersed human population[J]. Am. J. Hum. Genet., 2001,68:990-1018. |
| [1] | Luyao LI, Ning LYU, Yongjie ZHAI, Run ZHANG, Anxin YAN, Fuquan JIA. Genetic Polymorphism Analysis on Short Tandem Repeats of Y Chromosome in Han Population of Hohhot [J]. Current Biotechnology, 2025, 15(3): 544-554. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||