


生物技术进展 ›› 2022, Vol. 12 ›› Issue (6): 880-887.DOI: 10.19586/j.2095-2341.2022.0084
收稿日期:2022-05-24
接受日期:2022-06-30
出版日期:2022-11-25
发布日期:2022-11-30
通讯作者:
周晓今
作者简介:朱佳梦 E-mail: zhujiameng2021@163.com;
基金资助:
Jiameng ZHU1,2( ), Haiyang JIANG1, Rumei CHEN2, Xiaojin ZHOU2(
), Haiyang JIANG1, Rumei CHEN2, Xiaojin ZHOU2( )
)
Received:2022-05-24
Accepted:2022-06-30
Online:2022-11-25
Published:2022-11-30
Contact:
Xiaojin ZHOU
摘要:
凝胶阻滞实验(electrophoretic mobility shift assay,EMSA)是研究蛋白质与核酸结合的一种关键实验技术。EMSA技术兴起以来,使用放射性同位素、生物素标记核酸探针的手段已经非常成熟,但这两种传统的标记技术分别具有放射性探针稳定性差和生物素检测步骤复杂等缺点。近年来,尽管荧光标记探针逐渐被应用于EMSA中,但是对于利用荧光标记探针的EMSA仍缺乏系统的报道。对荧光标记的EMSA技术流程进行了优化和系统总结;利用6-羧基荧光素(6-carboxy-fluoroscine,FAM)标记ZmGRAS11启动子探针,通过EMSA检测其与Opaque2蛋白的结合,明确了蛋白和探针的适宜比例为8∶1。对GCN4 motif序列碱基进行突变并利用EMSA分析Opaque2与ZmGRAS11启动子之间的结合位点,结果表明GCN4 motif的“TGAC”核心基序在ZmGRAS11启动子与Opaque2蛋白的结合中可能起到了关键作用。研究结果为进一步探究Opaque2-ZmGRAS11转录调控模块在玉米籽粒发育中的作用机理提供了数据支撑。
中图分类号:
朱佳梦, 江海洋, 陈茹梅, 周晓今. 通过荧光标记的凝胶阻滞技术分析Opaque2蛋白与ZmGRAS11启动子的结合位点[J]. 生物技术进展, 2022, 12(6): 880-887.
Jiameng ZHU, Haiyang JIANG, Rumei CHEN, Xiaojin ZHOU. EMSA Experiments with Fluorescently Labeled Probes to Analyze the Binding Sites of Opaque2 Protein with the ZmGRAS11 Promoter[J]. Current Biotechnology, 2022, 12(6): 880-887.
| 试剂名称 | 配方 | pH |
|---|---|---|
| 1 mol·L-1 IPTG | 2.38301 g IPTG,10 mL 超纯水 | — |
| 裂解缓冲液 | 0.02 mol·L-1 咪唑,0.02 mol·L-1 Tris,0.5 mol·L-1 氯化钠,10% 甘油,0.5% TritonX-100 | 8.0 |
| 洗杂缓冲液A | 0.02 mol·L-1咪唑,0.02 mol·L-1 Tris,0.5 mol·L-1 氯化钠,10% 甘油,0.5% TritonX-100 | 8.0 |
| 洗杂缓冲液B | 0.04 mol·L-1咪唑,0.02 mol·L-1 Tris,0.5 mol·L-1 氯化钠,10%甘油,0.5% TritonX-100 | 8.0 |
| 洗杂缓冲液C | 0.08 mol·L-1 咪唑,0.02 mol·L-1 Tris,0.5 mol·L-1 氯化钠,10% 甘油,0.5% TritonX-100 | 8.0 |
| 洗脱缓冲液 | 0.25 mol·L-1咪唑,0.02 mol·L-1 Tris,0.5 mol·L-1 氯化钠,10% 甘油 | 8.0 |
| 1×置换缓冲液 | 0.05 mol·L-1 Tris,0.25 mol·L-1 氯化钠,0.005 mol·L-1 DTT | 7.5 |
| 5×TBE缓冲液 | 0.45 mol·L-1 Tris,0.45 mol·L-1 硼酸,0.01 mol·L-1 EDTA | 8.3 |
表1 实验所用到的试剂配方
Table 1 Formulation of reagent used in this study
| 试剂名称 | 配方 | pH |
|---|---|---|
| 1 mol·L-1 IPTG | 2.38301 g IPTG,10 mL 超纯水 | — |
| 裂解缓冲液 | 0.02 mol·L-1 咪唑,0.02 mol·L-1 Tris,0.5 mol·L-1 氯化钠,10% 甘油,0.5% TritonX-100 | 8.0 |
| 洗杂缓冲液A | 0.02 mol·L-1咪唑,0.02 mol·L-1 Tris,0.5 mol·L-1 氯化钠,10% 甘油,0.5% TritonX-100 | 8.0 |
| 洗杂缓冲液B | 0.04 mol·L-1咪唑,0.02 mol·L-1 Tris,0.5 mol·L-1 氯化钠,10%甘油,0.5% TritonX-100 | 8.0 |
| 洗杂缓冲液C | 0.08 mol·L-1 咪唑,0.02 mol·L-1 Tris,0.5 mol·L-1 氯化钠,10% 甘油,0.5% TritonX-100 | 8.0 |
| 洗脱缓冲液 | 0.25 mol·L-1咪唑,0.02 mol·L-1 Tris,0.5 mol·L-1 氯化钠,10% 甘油 | 8.0 |
| 1×置换缓冲液 | 0.05 mol·L-1 Tris,0.25 mol·L-1 氯化钠,0.005 mol·L-1 DTT | 7.5 |
| 5×TBE缓冲液 | 0.45 mol·L-1 Tris,0.45 mol·L-1 硼酸,0.01 mol·L-1 EDTA | 8.3 |
| 试管号 | 菌液加入量/mL | IPTG加入量/μL | IPTG诱导浓度/(mmol·L-1) |
|---|---|---|---|
| a | 6 | 0 | 0 |
| b | 6 | 1.2 | 0.2 |
| c | 6 | 2.4 | 0.4 |
| d | 6 | 3.6 | 0.6 |
| e | 6 | 4.8 | 0.8 |
| f | 6 | 6.0 | 1.0 |
表2 IPTG诱导浓度的优化条件
Table 2 Optimization of IPTG concentration for recombinant protein expression
| 试管号 | 菌液加入量/mL | IPTG加入量/μL | IPTG诱导浓度/(mmol·L-1) |
|---|---|---|---|
| a | 6 | 0 | 0 |
| b | 6 | 1.2 | 0.2 |
| c | 6 | 2.4 | 0.4 |
| d | 6 | 3.6 | 0.6 |
| e | 6 | 4.8 | 0.8 |
| f | 6 | 6.0 | 1.0 |
| 探针名称 | 序列(5'→3') | 标记 |
|---|---|---|
| 5'-GGCACTAGTCATGCTAGCTTGTGACTCATCCAACAAACAATAGTTATCC-3' | 5'-FAM | |
| FAM-R | 5'-GGATAACTATTGTTTGTTGGATGAGTCACAAGCTAGCATGACTAGTGCC-3' | 5'-FAM |
| WT-F | 5'-GGCACTAGTCATGCTAGCTTGTGACTCATCCAACAAACAATAGTTATCC-3' | — |
| WT-F | 5'-GGATAACTATTGTTTGTTGGATGAGTCACAAGCTAGCATGACTAGTGCC-3' | — |
| M1-F | 5'-GGCACTAGTCATGCTAGCTTGTCCAACAAACAATAGTTATCC-3' | — |
| M1-R | 5'-GGATAACTATTGTTTGTTGGACAAGCTAGCATGACTAGTGCC-3' | — |
| M2-F | 5'-GGCACTAGTCATGCTAGCTTGTTTTTTTTCCAACAAACAATAGTTATCC-3' | — |
| M2-R | 5'-GGATAACTATTGTTTGTTGGAAAAAAAACAAGCTAGCATGACTAGTGCC-3' | — |
| M3-F | 5'-GGCACTAGTCATGCTAGCTTGGGGGGGGTCCAACAAACAATAGTTATCC-3' | — |
| M3-R | 5'-GGATAACTATTGTTTGTTGGACCCCCCCCAAGCTAGCATGACTAGTGCC-3' | — |
| M4-F | 5'-GGCACTAGTCATGCTAGCTTGCAGCTCATCCAACAAACAATAGTTATCC-3' | — |
| M4-R | 5'-GGATAACTATTGTTTGTTGGATGAGCTGCAAGCTAGCATGACTAGTGCC-3' | — |
| M5-F | 5'-GGCACTAGTCATGCTAGCTTGTGGTCCATCCAACAAACAATAGTTATCC-3' | — |
| M5-R | 5'-GGATAACTATTGTTTGTTGGATGGACCACAAGCTAGCATGACTAGTGCC-3' | — |
| M6-F | 5'-GGCACTAGTCATGCTAGCTTGTGACCTGTCCAACAAACAATAGTTATCC-3' | — |
| M6-R | 5'-GGATAACTATTGTTTGTTGGACAGGTCACAAGCTAGCATGACTAGTGC-3' | — |
表3 ZmGRAS11探针聚合所用引物
Table 3 Probes in the ZmGRAS11 promoter
| 探针名称 | 序列(5'→3') | 标记 |
|---|---|---|
| 5'-GGCACTAGTCATGCTAGCTTGTGACTCATCCAACAAACAATAGTTATCC-3' | 5'-FAM | |
| FAM-R | 5'-GGATAACTATTGTTTGTTGGATGAGTCACAAGCTAGCATGACTAGTGCC-3' | 5'-FAM |
| WT-F | 5'-GGCACTAGTCATGCTAGCTTGTGACTCATCCAACAAACAATAGTTATCC-3' | — |
| WT-F | 5'-GGATAACTATTGTTTGTTGGATGAGTCACAAGCTAGCATGACTAGTGCC-3' | — |
| M1-F | 5'-GGCACTAGTCATGCTAGCTTGTCCAACAAACAATAGTTATCC-3' | — |
| M1-R | 5'-GGATAACTATTGTTTGTTGGACAAGCTAGCATGACTAGTGCC-3' | — |
| M2-F | 5'-GGCACTAGTCATGCTAGCTTGTTTTTTTTCCAACAAACAATAGTTATCC-3' | — |
| M2-R | 5'-GGATAACTATTGTTTGTTGGAAAAAAAACAAGCTAGCATGACTAGTGCC-3' | — |
| M3-F | 5'-GGCACTAGTCATGCTAGCTTGGGGGGGGTCCAACAAACAATAGTTATCC-3' | — |
| M3-R | 5'-GGATAACTATTGTTTGTTGGACCCCCCCCAAGCTAGCATGACTAGTGCC-3' | — |
| M4-F | 5'-GGCACTAGTCATGCTAGCTTGCAGCTCATCCAACAAACAATAGTTATCC-3' | — |
| M4-R | 5'-GGATAACTATTGTTTGTTGGATGAGCTGCAAGCTAGCATGACTAGTGCC-3' | — |
| M5-F | 5'-GGCACTAGTCATGCTAGCTTGTGGTCCATCCAACAAACAATAGTTATCC-3' | — |
| M5-R | 5'-GGATAACTATTGTTTGTTGGATGGACCACAAGCTAGCATGACTAGTGCC-3' | — |
| M6-F | 5'-GGCACTAGTCATGCTAGCTTGTGACCTGTCCAACAAACAATAGTTATCC-3' | — |
| M6-R | 5'-GGATAACTATTGTTTGTTGGACAGGTCACAAGCTAGCATGACTAGTGC-3' | — |
| 各个组分加入量/(μL) | 泳道 | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| 超纯水 | 14.0 | 13.5 | 13.0 | 12.0 | 10.0 | 8.0 | 6.0 | 4.0 |
| 5×结合缓冲液 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| O2蛋白 | 0 | 0.5 | 1.0 | 2.0 | 4.0 | 6.0 | 8.0 | 10.0 |
表4 不同蛋白与探针的比例设置
Table 4 Different treatments of protein to probe ratio
| 各个组分加入量/(μL) | 泳道 | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| 超纯水 | 14.0 | 13.5 | 13.0 | 12.0 | 10.0 | 8.0 | 6.0 | 4.0 |
| 5×结合缓冲液 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| O2蛋白 | 0 | 0.5 | 1.0 | 2.0 | 4.0 | 6.0 | 8.0 | 10.0 |
| 各个组分加入量/(μL) | 泳道 | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| 超纯水 | 14 | 10 | 5 | 0 | 0 | 0 | 0 | 0 |
| 5×结合缓冲液 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| O2蛋白 | 0 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| 竞争探针 | 0 | 0 | 5 | 10 | 10 | 10 | 10 | 10 |
表5 不同突变方式的竞争探针体系
Table 5 Reaction system of competitive probes with different mutations
| 各个组分加入量/(μL) | 泳道 | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| 超纯水 | 14 | 10 | 5 | 0 | 0 | 0 | 0 | 0 |
| 5×结合缓冲液 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| O2蛋白 | 0 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| 竞争探针 | 0 | 0 | 5 | 10 | 10 | 10 | 10 | 10 |
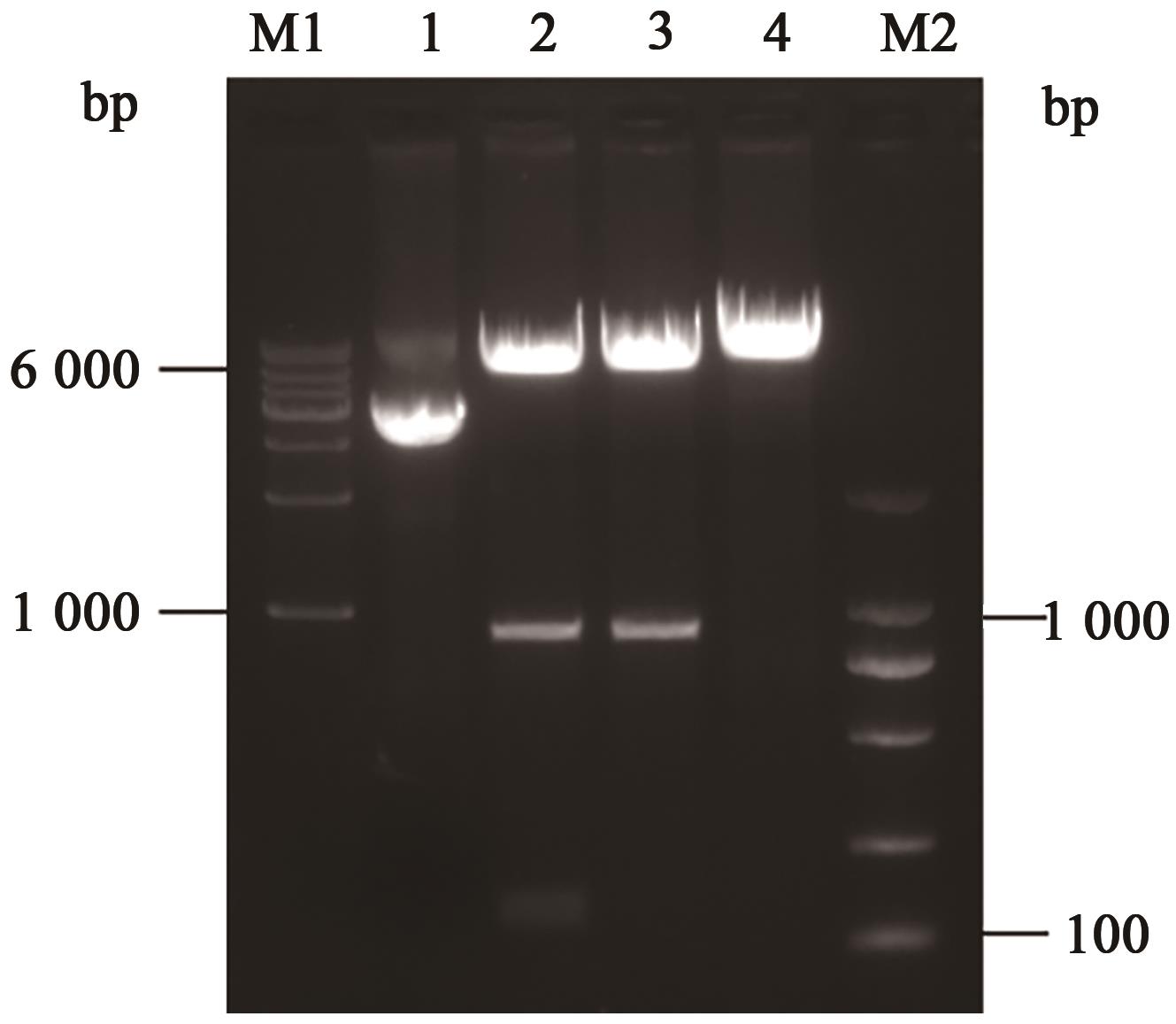
图2 pET28a-O2酶切鉴定电泳图注:M1—1 000 bp DNA ladder;M2—100 bp DNA ladder;泳道1—环状质粒;泳道2—EcoRⅠ、Xba Ⅰ双酶切;泳道3—EcoRⅠ单酶切;泳道4—Xba Ⅰ单酶切。
Fig. 2 Enzyme digestion identification of pET28a-O2
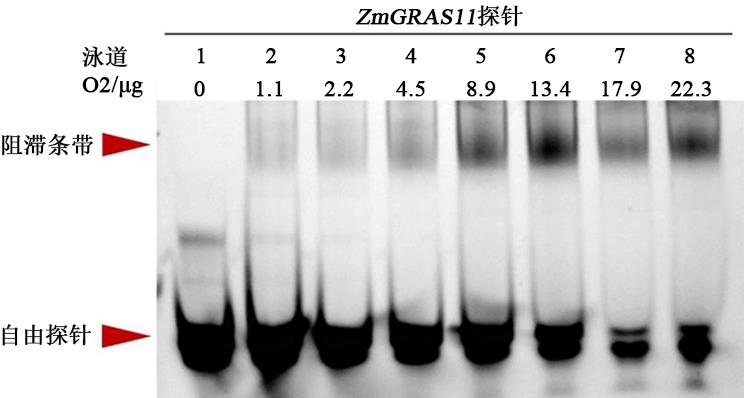
图4 EMSA检测不同浓度O2蛋白与ZmGRAS11探针的结合注:ZmGRAS11 探针为FAM荧光标记,各个泳道加入量均为20 pmol;泳道1~8中O2蛋白上样量分别为0、1.1、2.2、4.5、8.9、13.4、17.9、22.3 μg。
Fig. 4 The binding of O2 protein with ZmGRAS11 probe was detected by EMSA
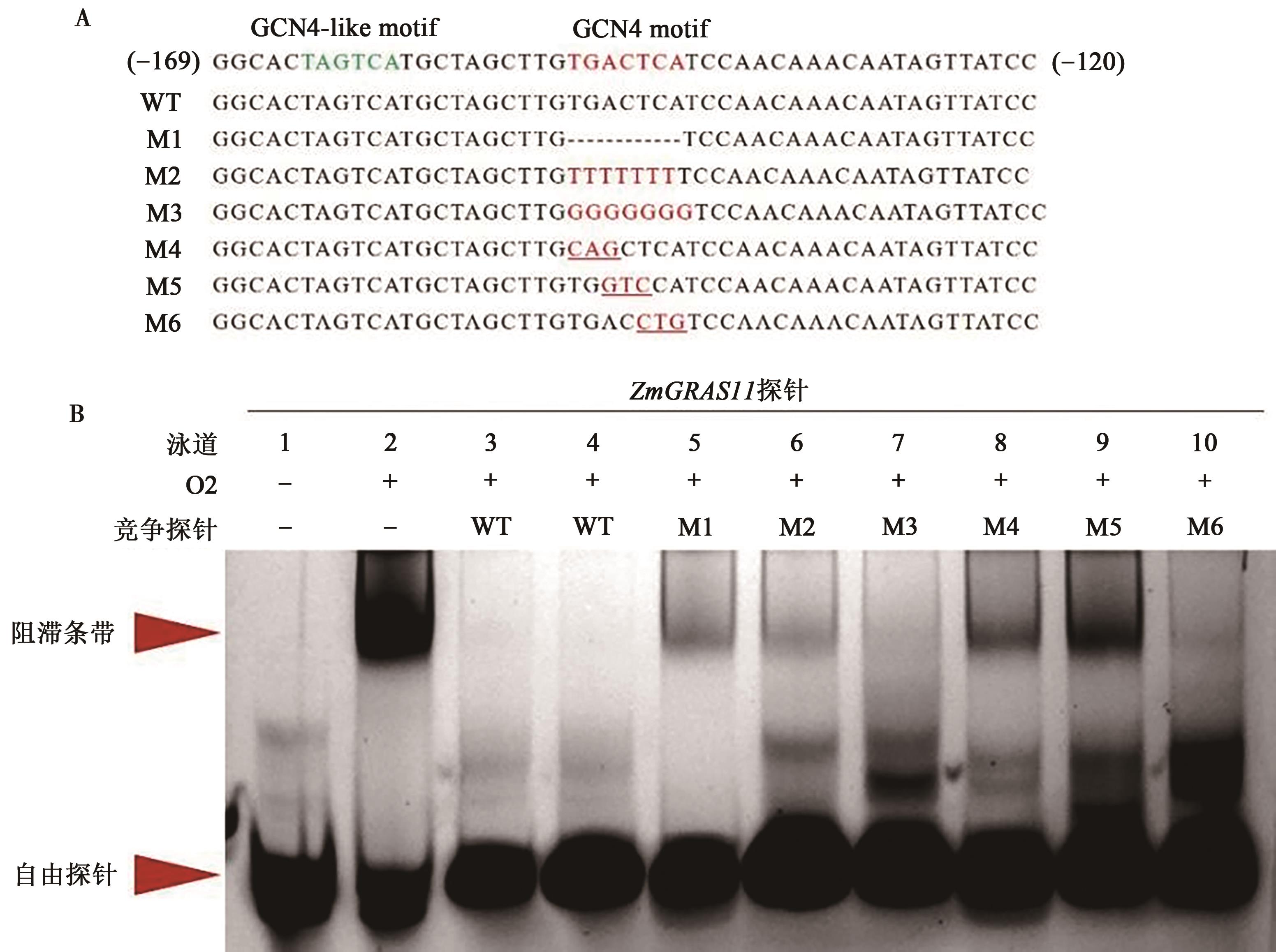
图5 不同方式突变对ZmGRAS11探针和O2蛋白结合力的影响A: ZmGRAS11启动子区探针设计;B: ZmGRAS11探针和O2蛋白结合力;ZmGRAS11 探针为FAM荧光标记,各个泳道加入量均为20 pmol;泳道2~10中O2蛋白的上样量均为8.9 μg。
Fig. 5 Effects of different mutations on the interaction between the ZmGRAS11-promorerprobe and O2 protein
| 1 | SITARAMAYYA A, WRIGHT L S, SIIEGEL F L. Enzymatic methylation of calmodulin in rat brain cytosol[J]. Biol. Chem., 1980, 255(18): 8894-8900. |
| 2 | HOWE C L, MOOSEKER M S, GRAVES T A. Brush-border calmodulin. A major component of the isolated microvillus core[J]. J.Cell Biol., 1980, 85: 916-923. |
| 3 | CHELM B K, GEIDUSCHEK E P. Gel electrophoretic separation of transcription complexes: an assay for RNA polymerase selectivity and a method for promoter mapping[J]. Nucl. Acids Res., 1979, 7(7): 1851-1867. |
| 4 | VARSHAVSKY A J, BAKAYEV V V, GEORGIEV G P. Heterogeneity of chromatin subunits in vitro and location of histone H1[J]. Nucl. Acids Res., 1976, 3(2): 477-492. |
| 5 | RAMANATHAN M, PORTER D F, KHAVARI P A. Methods to study RNA-protein interactions[J]. Nat. Methods., 2019, 16(3): 225-234. |
| 6 | REAM J A, LEWIS L K, LEWIS K A. Rapid agarose gel electrophoretic mobility shift assay for quantitating protein: RNA interactions[J]. Anal. Biochem., 2016, 511: 36-41. |
| 7 | SEO M, LEI L, EGLI M. Label-free electrophoretic mobility shift assay (EMSA) for measuring dissociation constants of protein-RNA complexes[J/OL]. Curr. Protoc. Nucl. Acid Chem., 2019, 76(1): e70[2022-06-01]. . |
| 8 | MAXAM A, GILBERT W S. A new method for sequencing DNA[J]. Proc. Natl. Acad. Sci. USA, 1977, 74: 560-565. |
| 9 | HELLMAN L M, FRIED M G. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions[J]. Nat. Protoc., 2007, 2(8): 1849-1861. |
| 10 | TIAN J, WANG C, XIA J, et al.. Teosinte ligule allele narrows plant architecture and enhances high-density maize yields[J]. Science, 2019, 365(6454): 658-664. |
| 11 | KONG D, PAN X, JING Y, et al.. ZmSPL10/14/26 are required for epidermal hair cell fate specification on maize leaf[J]. New Phytol., 2021, 230(4): 1533-1549. |
| 12 | XIE Y, LIU Y, WANG H, et al.. Phytochrome-interacting factors directly suppress MIR156 expression to enhance shade-avoidance syndrome in Arabidopsis [J]. Nat. Commun., 2017, 8(1): 1-11. |
| 13 | LI C, SONG R. The regulation of zein biosynthesis in maize endosperm[J]. Theor. Appl. Genet., 2020, 133(5): 1443-1453. |
| 14 | LI C, YUE Y, CHEN H, et al.. The ZmbZIP22 transcription factor regulates 27-kD γ-Zein gene transcription during maize endosperm development[J]. Plant Cell, 2018, 30(10): 2402-2424. |
| 15 | QIAO Z, QI W, WANG Q, et al.. ZmMADS47 regulates Zein gene transcription through interaction with Opaque2[J/OL]. PLoS Genet., 2016, 12(4): e1005991[2022-06-01]. . |
| 16 | VICENTE-CARBAJOSA J, MOOSE S P, PARSONS R L, et al.. A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2[J]. Proc. Natl. Acad. Sci. USA, 1997, 94(14): 7685-7690. |
| 17 | DENG Y, WANG J, ZHANG Z, et al.. Transactivation of Sus1 and Sus2 by Opaque2 is an essential supplement to sucrose synthase-mediated endosperm filling in maize[J]. Plant Biotechnol. J., 2020, 18(9): 1897-1907. |
| 18 | ZHOU Z, SONG L, ZHANG X, et al.. Introgression of opaque2 into waxy maize causes extensive biochemical and proteomic changes in endosperm[J/OL]. PLoS ONE, 2016, 11(7): e0158971[2022-06-01]. . |
| 19 | LI Y, MA S, ZHAO Q, et al.. ZmGRAS11, transactivated by Opaque2, positively regulates kernel size in maize[J]. J. Integr. Plant Biol., 2021, 63(12): 2031-2037. |
| 20 | GAO Y, AN K, GUO W, et al.. The endosperm-specific transcription factor TaNAC019 regulates glutenin and starch accumulation and its elite allele improves wheat grain quality[J]. Plant Cell, 2021, 33(3): 603-622. |
| 21 | LI C, QIAO Z, QI W, et al.. Genome-wide characterization of cis-acting DNA targets reveals the transcriptional regulatory framework of opaque2 in maize[J]. Plant Cell, 2015, 27(3): 532-545. |
| 22 | LIU Y, MA M, LI G, et al.. Transcription factors FHY3 and FAR1 regulate light-induced CIRCADIAN CLOCK ASSOCIATED1 gene expression in Arabidopsis [J]. Plant Cell, 2020, 32(5): 1464-1478. |
| 23 | ZONG W, TANG N, YANG J, et al.. Feedback regulation of ABA signaling and biosynthesis by a bZIP transcription factor targets drought-resistance-related genes[J]. Plant Physiol., 2016, 171(4): 2810-2825. |
| 24 | 刘蕾,周欣悦,朱桢,等. 副溶血弧菌外膜铁蛋白受体pvuA基因的原核表达及产物的诱导条件优化[J].生物技术进展, 2022,12(3):396-404. |
| 25 | 陈威风,崔丽伟,常惟丹,等.金黄色葡萄球菌肠毒素A、B、C、D、E重组载体的构建及表达[J].生物技术进展, 2022,12(2):313-317. |
| 26 | GALHANO R, ILLANA A, RYDER L S, et al.. Tpc1 is an important Zn(II)2Cys6 transcriptional regulator required for polarized growth and virulence in the rice blast fungus[J/OL]. PLoS Pathog., 2017, 13(7): e1006516[2022-06-01]. . |
| 27 | ZHANG Z, ZHENG X, YANG J, et al.. Maize endosperm-specific transcription factors O2 and PBF network the regulation of protein and starch synthesis[J]. Proc. Natl. Acad. Sci. USA, 2016, 113(39): 10842-10847. |
| 28 | SHPJI T, HASHIMOTO T. Tobacco MYC2 regulates jasmonate-inducible nicotine biosynthesis genes directly and by way of the NIC2-locus ERF genes[J]. Plant Cell Physiol., 2011, 52(6): 1117-1130. |
| 29 | MENG Y, WANG Z, WANG Y, et al.. The MYB activator WHITE PETAL1 associates with MtTT8 and MtWD40-1 to regulate carotenoid-derived flower pigmentation in Medicago truncatula [J]. Plant Cell, 2019, 31(11): 2751-2767. |
| 30 | JI C, XU L, LI Y, et al.. The O2-ZmGRAS11 transcriptional regulatory network orchestrates the coordination of endosperm cell expansion and grain filling in maize[J]. Mol. Plant., 2022, 15(3): 468-487. |
| [1] | 徐振华, 高世伟, 高大伟, 于艳敏, 刘海英, 武洪涛, 张书利, 孙忠义, 王昕, 闫平. 生物育种科技创新助力种业发展及展望[J]. 生物技术进展, 2025, 15(4): 557-564. |
| [2] | 吕婉婉, 郑林, 庞红盈, 高宏波, 王宏芝. 杨树根蘖苗发生机理研究进展[J]. 生物技术进展, 2025, 15(3): 372-379. |
| [3] | 刘卓颖, 周晓今, 黄燕丽, 逄森. 玉米盐胁迫和MeJA处理下的转录组联合分析[J]. 生物技术进展, 2025, 15(2): 263-275. |
| [4] | 张兰兰, 李才华, 方雨竹, 宋岩, 康婉琳, 李志宇, 张晓, 张锐. 线粒体SSR分子标记在植物中的应用进展[J]. 生物技术进展, 2023, 13(6): 821-826. |
| [5] | 李敏, 王磊, 邹俊杰. 我国转基因抗虫耐除草剂玉米产业化应用面临的机遇与挑战[J]. 生物技术进展, 2023, 13(2): 157-165. |
| [6] | 孙卉, 张春义, 姜凌. 药用植物分子农场研究进展[J]. 生物技术进展, 2023, 13(1): 65-71. |
| [7] | 杨洋, 王凤林, 刘德, 罗园园, 朱建华. CRISPR⁃Cas9技术在植物次生代谢物生产中的研究进展[J]. 生物技术进展, 2022, 12(6): 806-816. |
| [8] | 乔朝辉, 陈亮. RNA聚合酶Ⅱ启动子近端暂停/释放的动态调控及生理作用研究进展[J]. 生物技术进展, 2022, 12(5): 705-710. |
| [9] | 周琴, 谢钰容. 光信号转导因子HY5调控拟南芥分枝的功能研究[J]. 生物技术进展, 2022, 12(3): 379-386. |
| [10] | 费云燕, 杨军, 景德道, 林添资, 李闯, 钱华飞, 曾生元, 韩华新, 龚红兵. CRISPR/Cas技术在抗除草剂作物育种中的研究与应用进展[J]. 生物技术进展, 2022, 12(2): 189-197. |
| [11] | 黄赳, 师双峰, 张二特, 李梅, 于跃, 刘昱辉. 矮珍珠硝酸盐转运蛋白基因GeNRT2.1的克隆和功能研究[J]. 生物技术进展, 2022, 12(2): 256-264. |
| [12] | 王欣, 张天柱. 园艺作物花青素合成调控研究进展[J]. 生物技术进展, 2022, 12(1): 10-16. |
| [13] | 许慧露, 徐岷, 张炜. 敲除集胞藻PCC 6803中编码乳酸脱氢酶的slr1556基因对生物合成乙醇的影响[J]. 生物技术进展, 2022, 12(1): 105-111. |
| [14] | 李梦杰, 贺晓云, 仝涛, 梁晋刚, 黄昆仑. 阿根廷转基因作物安全管理制度概况及进展[J]. 生物技术进展, 2021, 11(6): 676-687. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||